Single molecule biophysics in vitro
Current research:
We develop a high sensitivity single molecule force spectroscopy instrument to study the mechano-kinetic properties of biomolecules and their interactions.
Published projects:
→ Protein Folding
Full distance-resolved folding energy landscape of one single protein molecule
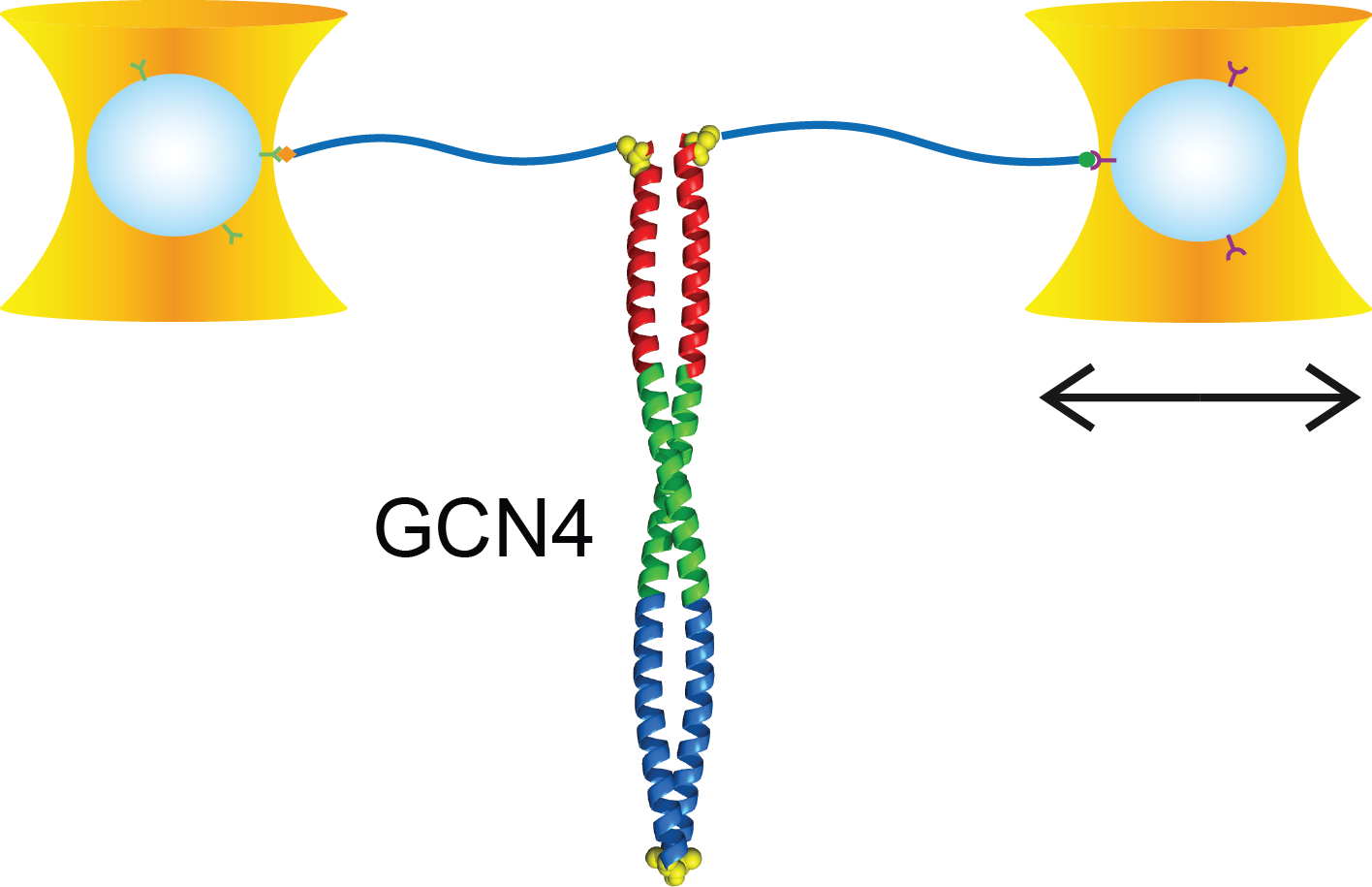
Kinetic bulk and single molecule folding experiments characterize barrier properties but the shape of folding landscapes between barrier top and native state is difficult to access. Here, we directly extract the full free energy landscape of a single molecule of the GCN4 leucine zipper using dual beam optical tweezers. To this end, we use deconvolution force spectroscopy to follow an individual molecule’s trajectory with high temporal and spatial resolution. We find a heterogeneous energy landscape of the GCN4 leucine zipper domain. The energy profile is divided into two stable C-terminal heptad repeats and two less stable repeats at the N-ter- minus. Energies and transition barrier positions were confirmed by single molecule kinetic analysis. We anticipate that deconvolution sampling is a powerful tool for the model-free investigation of protein energy landscapes.
doi: 10.1073/pnas.0909854107
→ Molecular motors
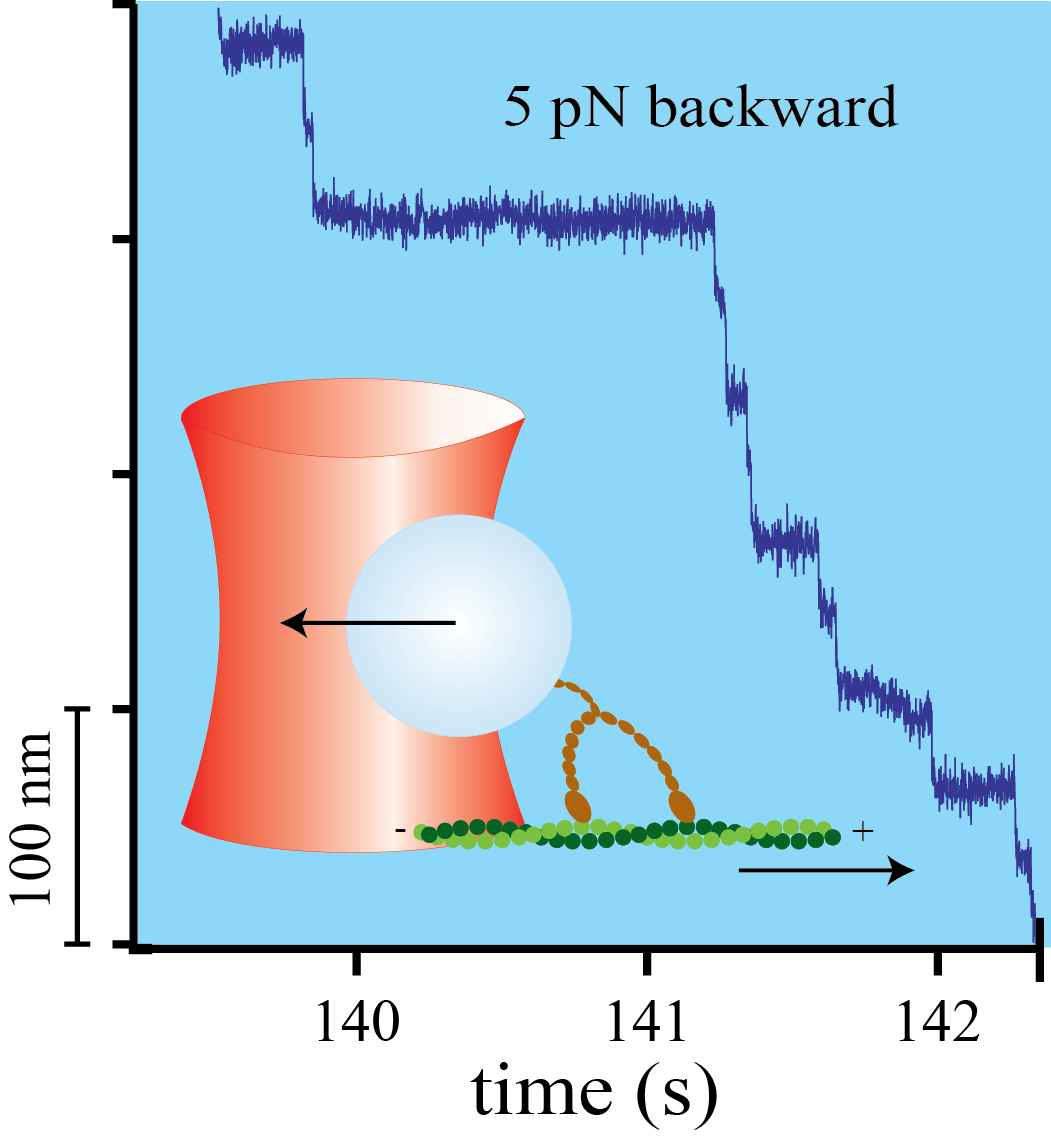
Mechanical asymmetry of the myosin-V - actin bond
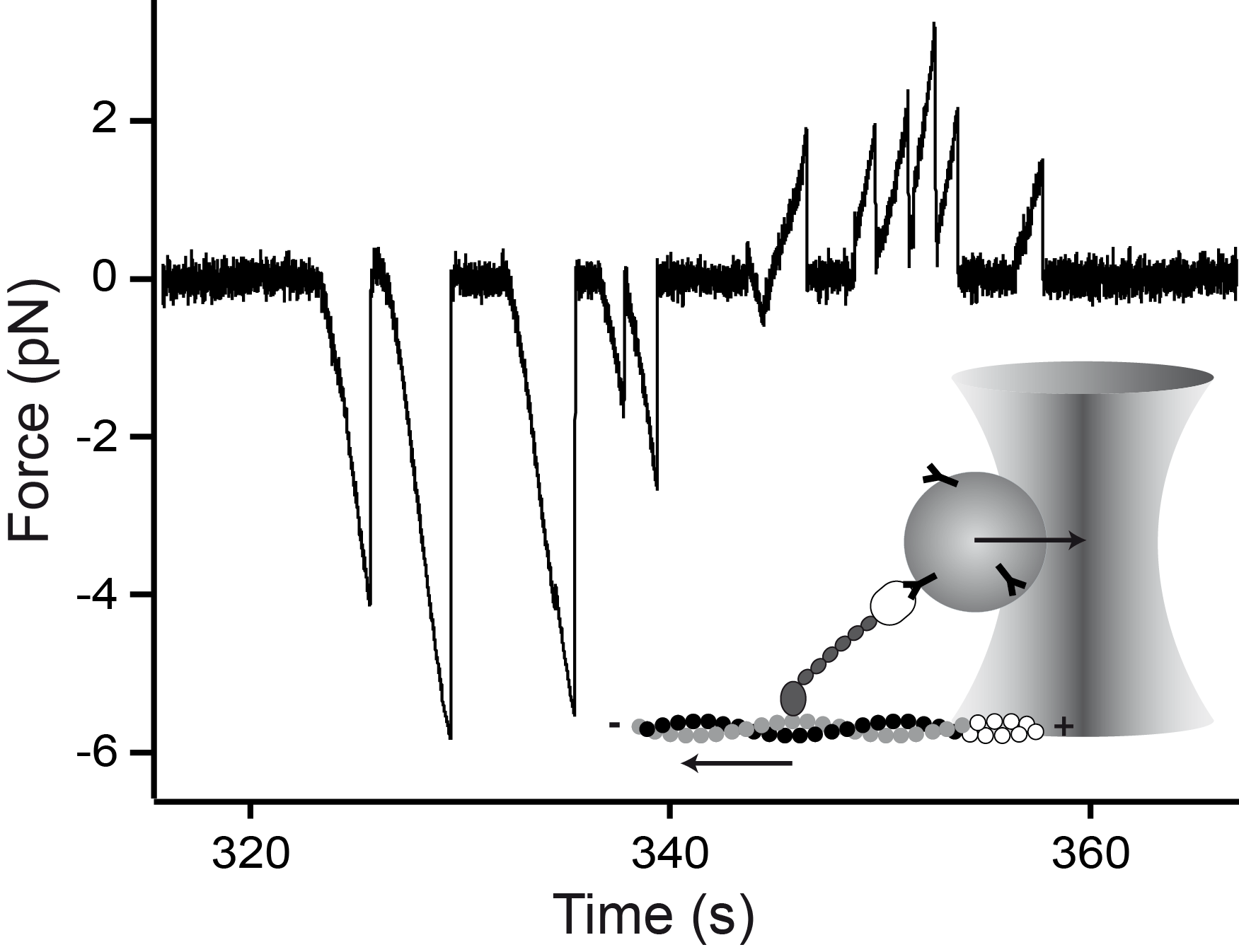
Myosin-V is a two-headed molecular motor taking multiple ATP-dependent steps toward the plus end (forward) of actin filaments. At high mechanical loads, the motor processively steps toward the minus end (backward) even in the absence of ATP, whereas analogous forward steps cannot be induced. The detailed mechanism underlying this mechanical asymmetry is not known. We investigate the effect of force on individual single headed myosin-V constructs bound to actin in the absence of ATP. If pulled forward, the myosin-V head dissociates at forces twice as high than if pulled backward. Moreover, backward but not forward distances to the unbinding barrier are dependent on the lever arm length. This asymmetry of unbinding force distributions in a single headed myosin forms the basis of the two-headed asymmetry. Under load, the lever arm functions as a true lever in a mechanical sense.
doi: 10.1016/j.bpj.2009.10.017
Myosin-V is a mechanical ratchet
Myosin-V is a linear molecular motor that hydrolyzes ATP to move processively toward the plus end of actin filaments. Motion of this motor under low forces has been studied recently in various single-molecule assays. In this paper we show that myosin-V reacts to high forces as a mechanical ratchet. High backward loads can induce rapid and processive backward steps along the actin filament. This motion is completely independent of ATP binding and hydrolysis. In contrast, forward forces cannot induce ATP-independent forward steps. We can explain this pronounced mechanical asymmetry by a model in which the strength of actin binding of a motor head is modulated by the lever arm conformation. Knowledge of the complete force–velocity dependence of molecular motors is important to understand their function in the cellular environment.
doi: 10.1073/pnas.0510191103