Ribonucleases
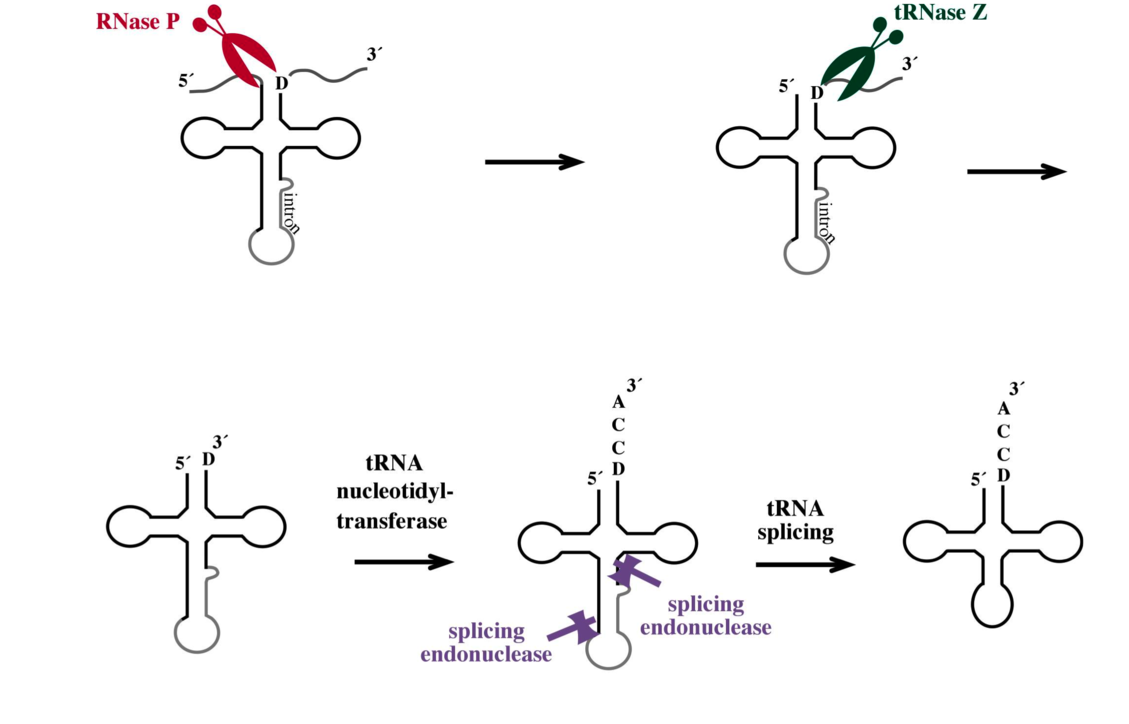
All RNA molecules are transcribed into precursor RNAs and have to undergo a variety of processing events to become functional RNAs. In addition, they also have to undergo controlled degradation. The generation and regulated turnover of these RNA molecules is catalysed by ribonucleases. Only few of these important enzymes have been identified in Archaea. Therefore we are investigating and identifying new ribonucleases and are characterising them using in vitro and in vivo methods in Haloarchaea.
We isolated the tRNA processing enzyme tRNase Z and are currently characterising this enzyme in detail. In addition, we investigate the mRNA 3'-end processing protein CPSF, the RNase G/E, the RppH protein and RNase J. To learn more about the evolution of these ribonucleases we compare them to their eukaryotic counterparts in yeast and plants.