Research in the Department of Molecular Genetics and Cell Biology
1. Polarity Establishment in Eucaryotic Cells
Establishment of cellular polarity is a key requisite for eucaryotic life. Misfunction in polarity establishment results in severe consequences for the cell and the organism including cell death or development of diseases. Many of the meachnisms underlying polarity establishment are conserved among eucaryotic cells. Due to it's ease in manipulation, we employ the budding yeast Saccharomyces cerevisiae as a model organism to study establishment and maintenance of cellular polarity.
The assembly of the bud site is a remarkable and hardly understood process. Kick-started by a local pulse of active Cdc42, hundreds of different proteins, lipids, and other macromolecules are attracted to a single spot of less than one micron. After 10 minutes the bud site starts to expand and to grow into a new daughter cell. The structure of the bud site is too complex and probably too amorphous to be assembled by a single linear, step-wise process. The convergence of parallel pathways that influence and inform each other represents an alternative, and probably more realistic view how the bud site forms and thus polarity establishes.
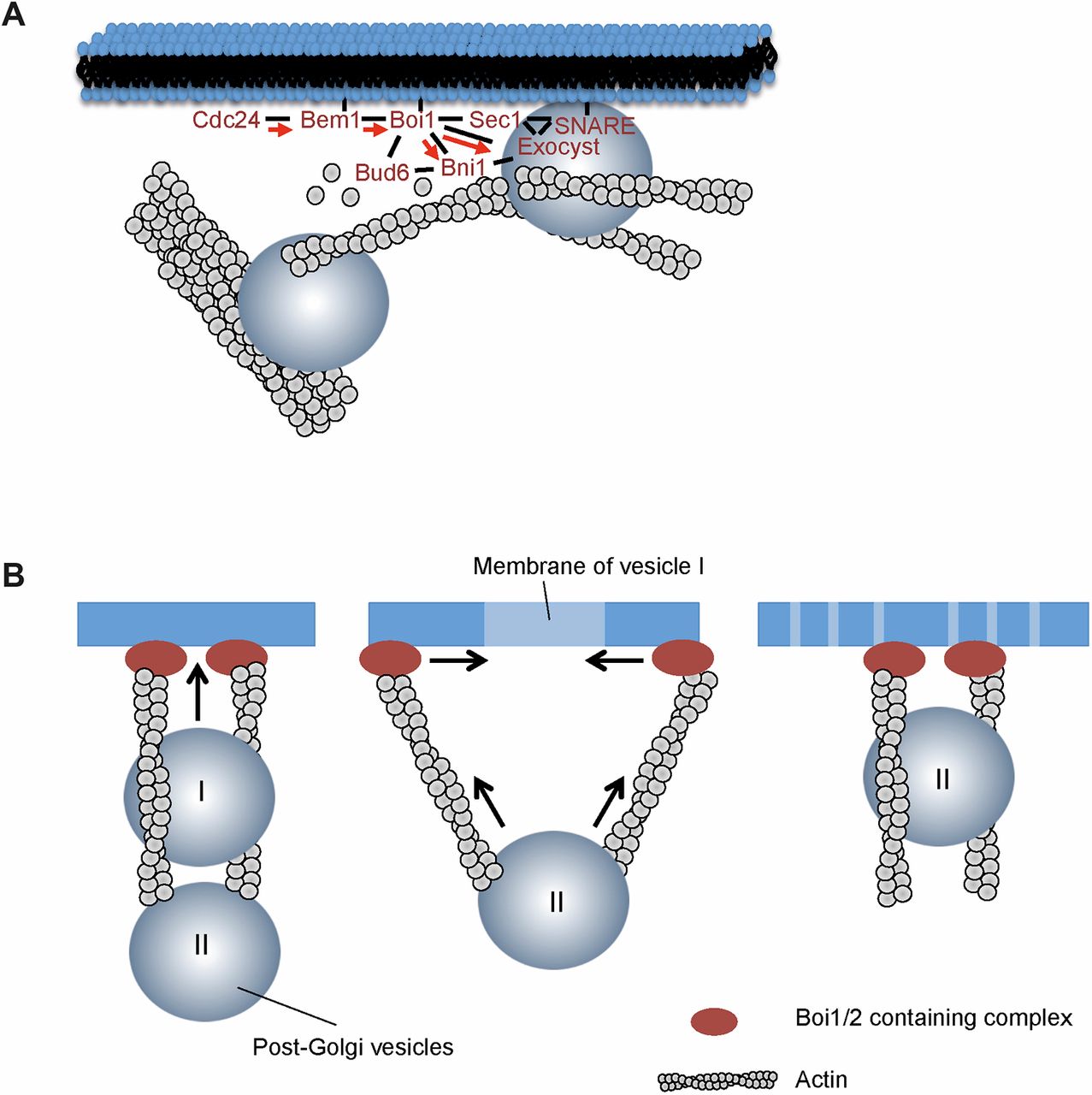
After bud site establishment, the yeast cells continue polar growth of the bud. This polar growth is the result of a conserved and complex interplay between RhoGTPase-based signalling, cytoskeletal organization, and vesicular traffic and fusion (figure 1.1).
We investigate the formation, composition and function of protein interaction networks that drive polarity establishment, polar growth and cytokinesis in vivo. Through temporal interaction analysis we are able to describe these networks as time- and localization-dependent conversions of different interaction states inside the living cell.
Experimental strategies in this topic include time resolved fluorescence microscopy, high throughput interaction screens, genetic manipulation of yeast and biochemical assays.
2. Cellular Biochemistry of the Septins
The septins were only discovered in the 1970s and represent a class of GTP binding cytoskeletal proteins sharing structural features of the Ras family of GTPases (Grupp & Gronemeyer, 2022). They form non-polar filaments that are indispensable for eukaryotic life. Dysfunction of septin subunit expression and filament formation is related to severe cellular phenotypes and to diseases such as cancer, male infertility, neurodevelopmental disorders and others.
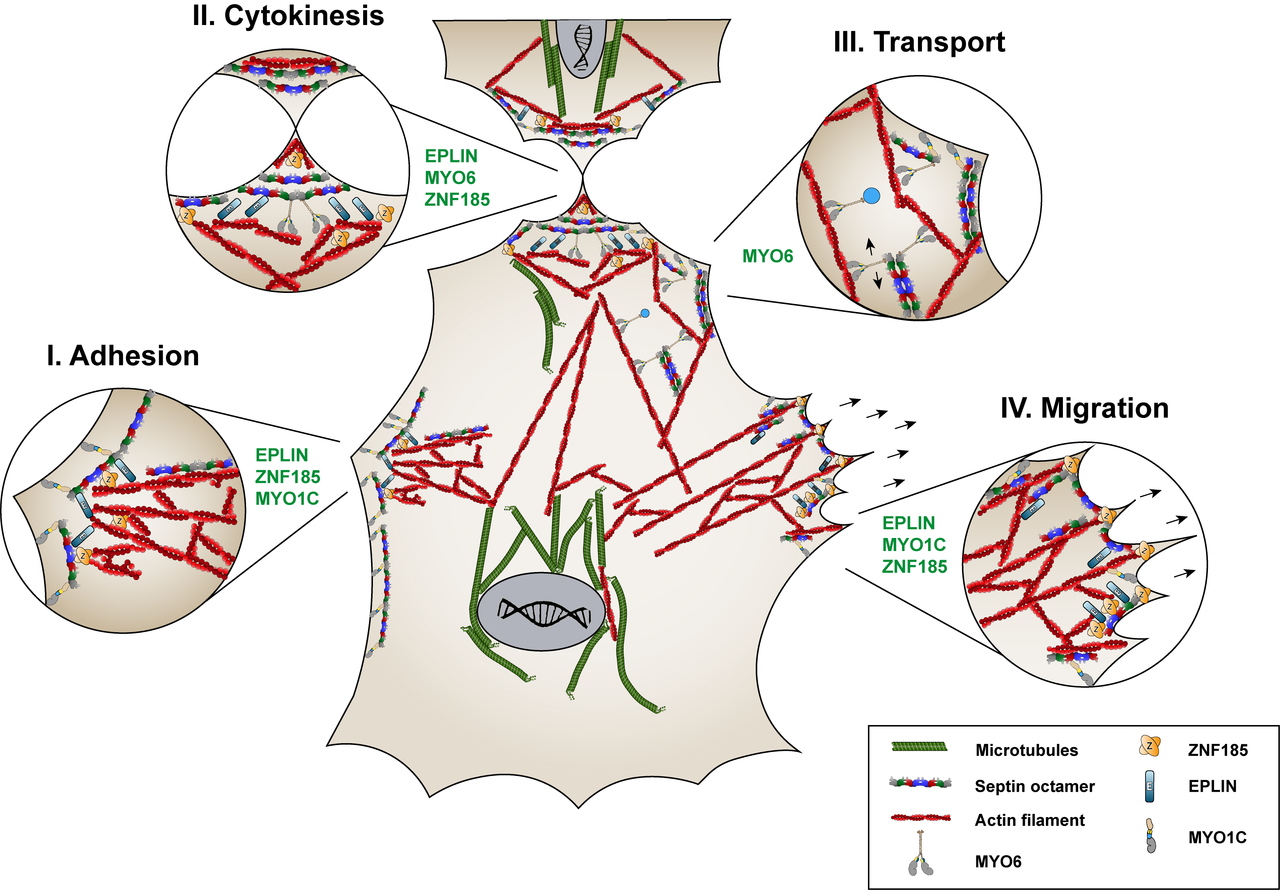
The human septin SEPT9 is related to several diseases. Identifying novel interaction partners of SEPT9 should help to gain insights into the intracellular processes leading to SEPT9 related cellular misfunctions and disease onset. Applying affinity purification of SEPT9 complexes combined with quantitative mass spectrometry, we have identified 42 interaction partners of SEPT9 (Hecht et al., 2019). Currently we are investigating the role of novel interaction partners with respect to their SEPT9 dependent function in cellular adhesions, cytokinesis and intracellular vesicle transport (figure 2.1).
In this project we apply siRNA mediated knock down of candidate genes, immunofluorescence- and live cell microscopy as well as in vitro assays using purified proteins.
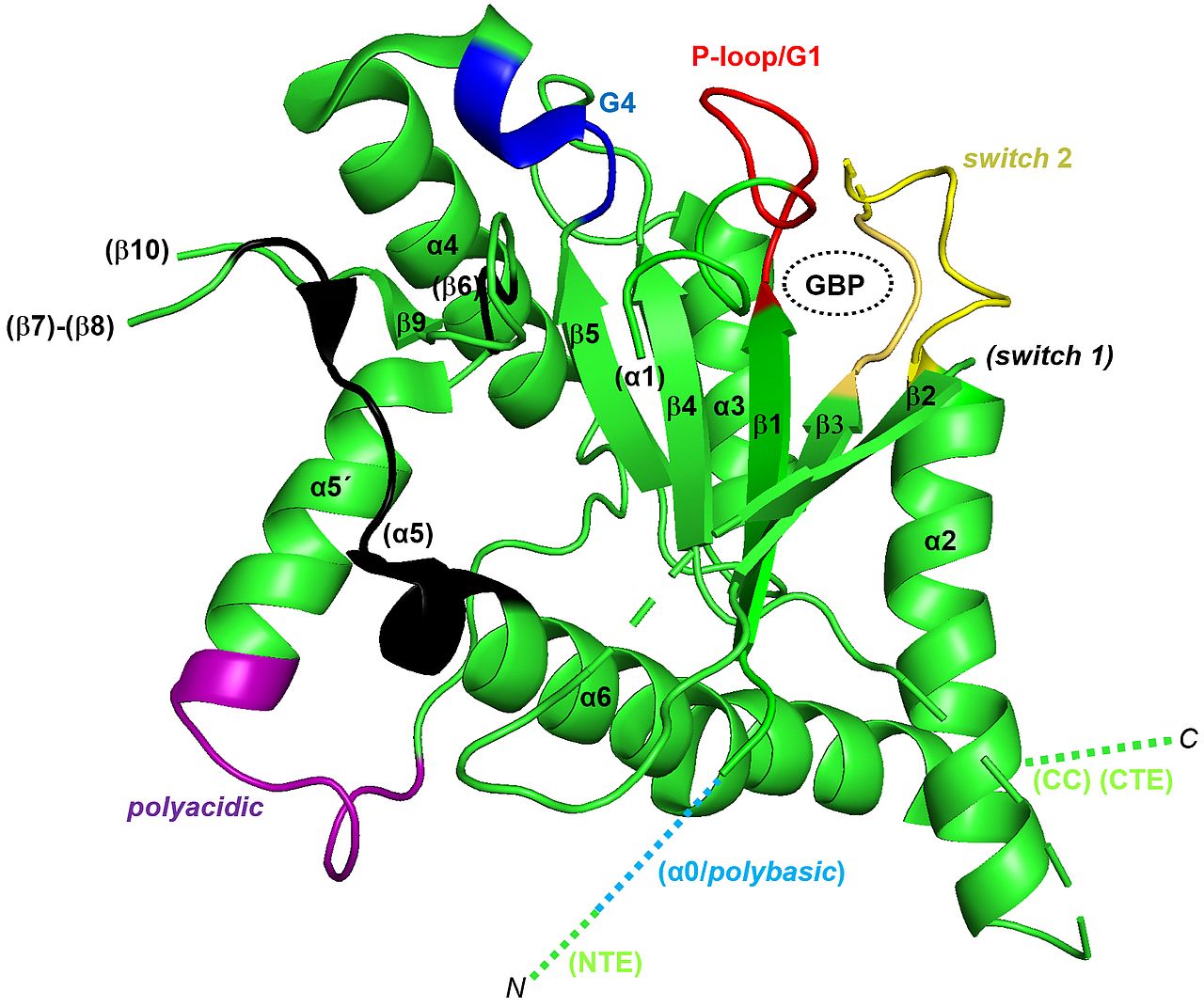
Septins were discovered in the baker’s yeast Saccharomyces cerevisiae and still today yeast serves as a prime model organism for research on septins due to its ease of genetic manipulation and the limited number of different septin subunits. We aim at understanding the mechanisms of septin filament formation and nucleotide binding by combining biochemical assays, structural biology and cell biology. Our laboratory presented the first crystal structure of a yeast septin (Brausemann et al. 2016) (figure 2.2) and the first crystal structure of a tetrameric yeast septin complex (Grupp et al. 2024). Currently we study the mechanisms of nucleotide binding and hydrolysis and the influence of these processes on septin protofilament formation.
Some questions, however, cannot be addressed in the wetlab since the required molecules are not available. To address these problems, we apply molecular dynamics based in silico approaches (Grupp et al. 2023).