Beneficial effectors - symbiont-guided plant protection
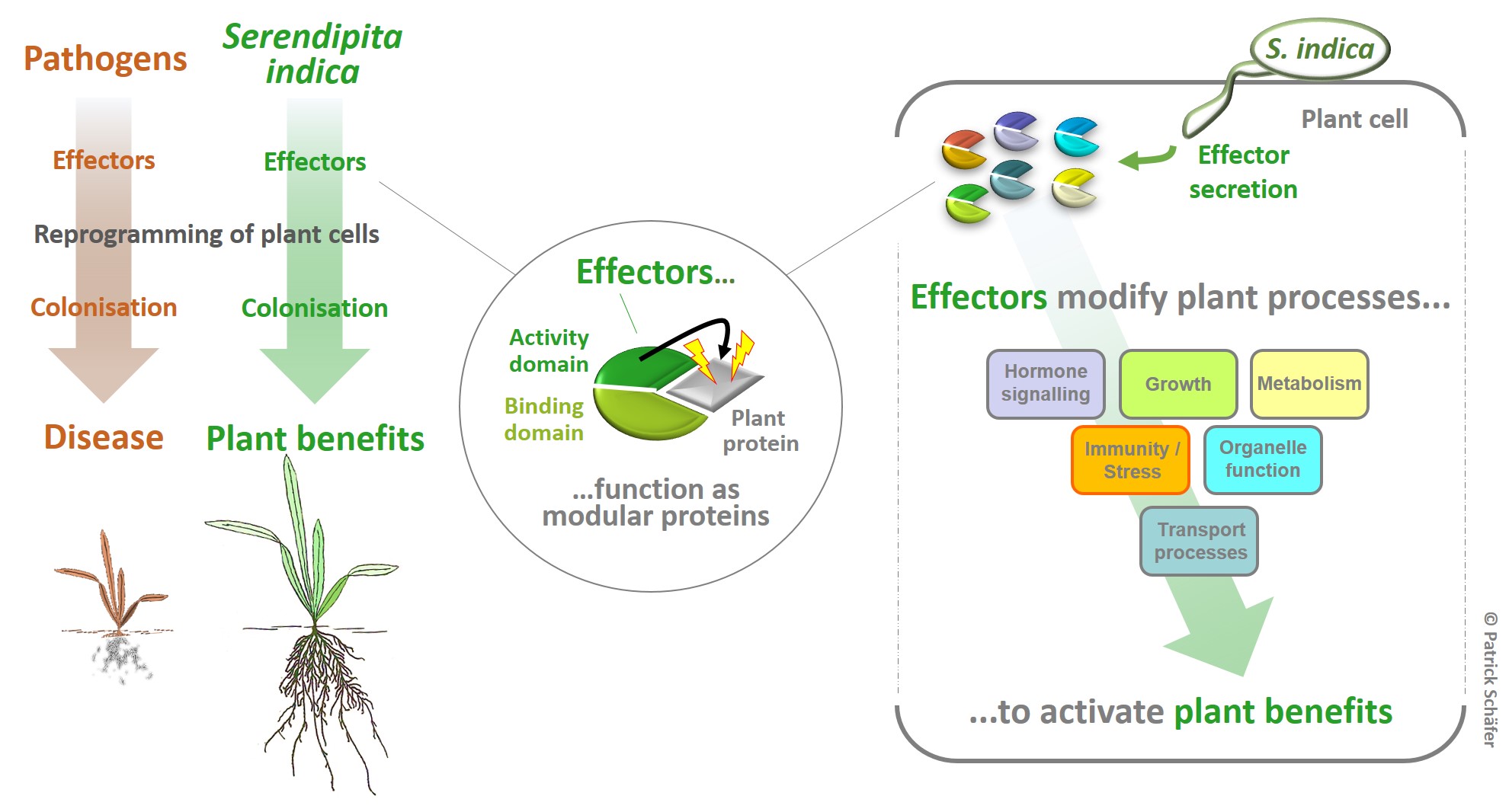
Background: Beneficial symbionts (mutualists) and pathogens secrete proteins (termed effectors) for the targeted reprogramming of host signalling as an essential step in colonising host plants. However, the outcome of their effector activities is obviously very different. While the activities of pathogen effectors result in disease outbreaks, effectors from beneficial symbionts might contribute to the beneficial effects observed in colonised plants. Since the mutualist Serendipita indica protects Arabidopsis and all major crops against environmental stress (i.e. drought, heat, pathogen attack) mutualistic S. indica effectors represent a currently untapped resource for sustainable improvement of stress resilience and growth under climate-associated stress in a broad range of plants.
Project: We have identified >100 effector candidatess in the genome of S. indica (S. indica effectors, SIEs) and developed a screening strategy to identify SIEs that enhance stress protection in plants. In addition to comprehensive yeast two-hybrid based protein network analyses (with 13K Arabidopsis genes), we phenotyped all SIEs for their specific function in changing distinct growth and stress pathways. Finally, all SIEs were expressed in whole plants and screened for improved growth, immunity, salt and drought stress. Based on these screens, we have identified SIEs with various beneficial activities including stress protection against climate stress in Arabidopsis. The aim is to apply SIE-derived activities to decipher the hierarchies and connection of stress signalling in plants. In biochemical, cell and structural biology-based studies, we further elucidate the mode of action of SIEs.
Lab tools / techniques: Protoplast-based phenotyping, protein network studies, biochemical analyses of abiotic/biotic stress assays, non-invasive cell imaging.
Relevant publications:
Li, T., Wang, Q., Feng, R., Fan, G., Li, W., Du, Y., Zhang, M., Schäfer, P., Tyler, B.M., Shan, W. (2019) Negative regulators of plant immunity derived from cinnamyl alcohol dehydrogenases are targeted by multiple Phytophthora Avr3a-like effectors. Accepted in New Phytologist doi.org/10.1111/nph.16139.
Patron, N., Orzaez, D., Marillonnet, S., Warzecha, H., Matthewman, C., Youles, M., Raitskin, O., Leveau, A., Farre-Martinez, G., Rogers, C., Smith, A., Hibberd, J., Webb, A., Locke, J., Schornack, S., Ajioka, J., Baulcombe, D., Zipfel, C., Kamoun, S., Jones, J., Kuhn, H., Robatzek, S., Van Esse, H.P., Oldroyd, G., Sanders, D., Martin, C., Field, R., O'Connor, S., Fox, S., Wulff, B., Miller, B., Breakspear, A., Radhakrishnan, G., Delaux, P.M., Loque, D., Granell, A., Tissier, A., Shih, P., Brutnell, T., Quick, P., Rischer, H., Fraser, P., Aharoni, A., Raines, C., South, P., Ané, J.M., Hamberger, B., Langdale, J., Stougaard, J., Bouwmeester, H., Udvardi, M., Murray, J., Ntoukakis, V., Schäfer, P., Denby, K., Edwards, K., Osbourn, A., Haseloff, J. (2015) Standards for Plant Synthetic Biology: A Common Syntax for Exchange of DNA Parts. New Phytologist 208:13-19.
Döhlemann, G., Requena, N., Schäfer, P., Brunner, F., O’Connell, R., Parker, J.E. (2014) Reprogramming of plant cells by filamentous plant-colonising microbes. New Phytologist 204:803-14.