Publications 2020
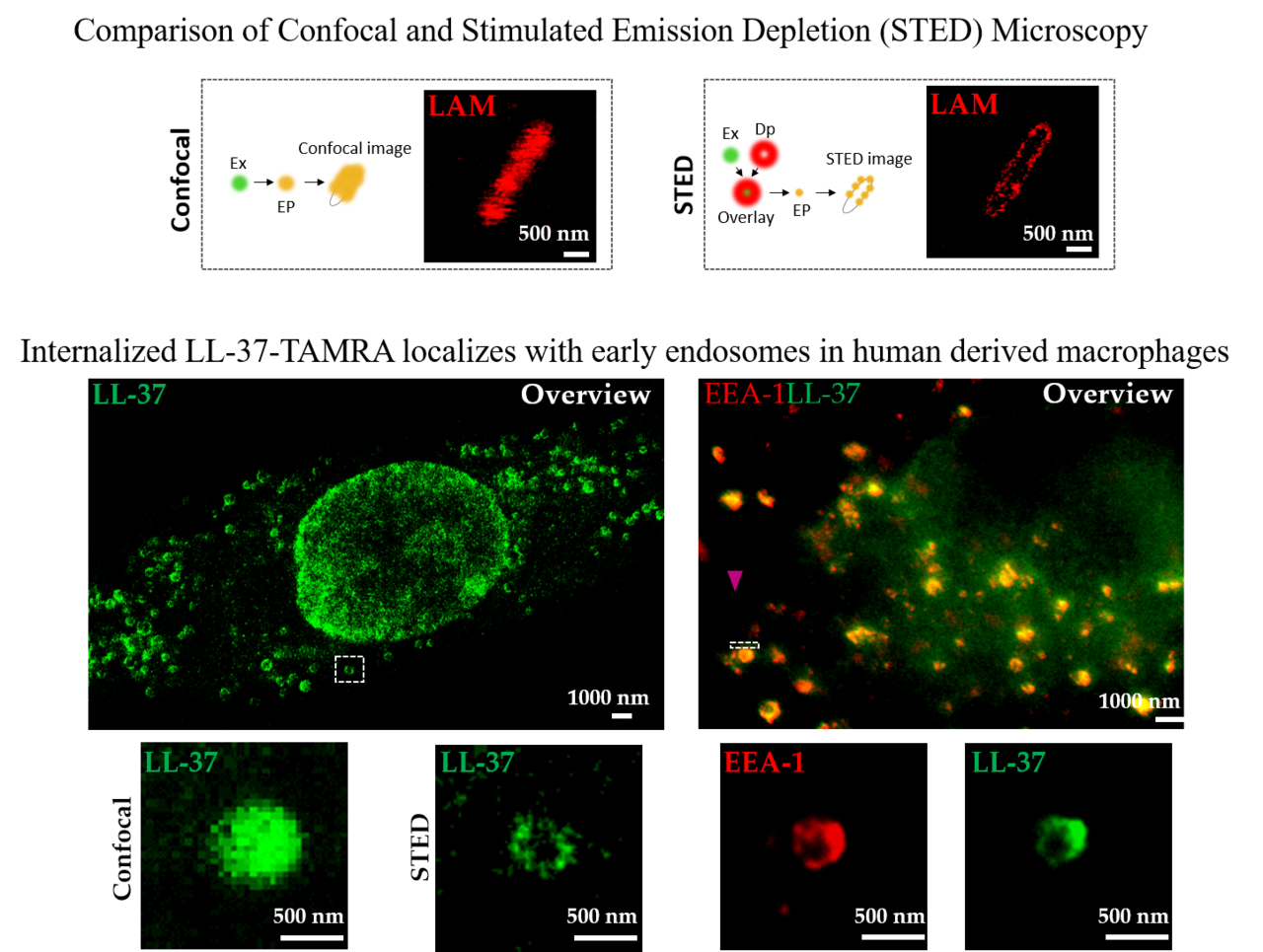
Super-Resolution Microscopy Reveals a Direct Interaction of Intracellular Mycobacterium tuberculosis with the Antimicrobial Peptide LL-37
Dhruva Deshpande, Mark Grieshober, Fanny Wondany, Fabian Gerbl, Reiner Noschka, Jens Michaelis and Steffen Stenger
The antimicrobial peptide LL-37 inhibits the growth of the major human pathogen Mycobacterium tuberculosis (Mtb), but the mechanism of the peptide–pathogen interaction inside human macrophages remains unclear. Super-resolution imaging techniques provide a novel opportunity to visualize these interactions on a molecular level. Here, we adapt the super-resolution technique of stimulated emission depletion (STED) microscopy to study the uptake, intracellular localization and interaction of LL-37 with macrophages and virulent Mtb. We demonstrate that LL-37 is internalized by both uninfected and Mtb infected primary human macrophages. The peptide localizes in the membrane of early endosomes and lysosomes, the compartment in which mycobacteria reside. Functionally, LL-37 disrupts the cell wall of intra- and extracellular Mtb, resulting in the killing of the pathogen. In conclusion, we introduce STED microscopy as an innovative and informative tool for studying host–pathogen–peptide interactions, clearly extending the possibilities of conventional confocal microscopy.
Int. J. Mol. Sci. 2020, 21(18), 6741
doi.org/10.3390/ijms21186741
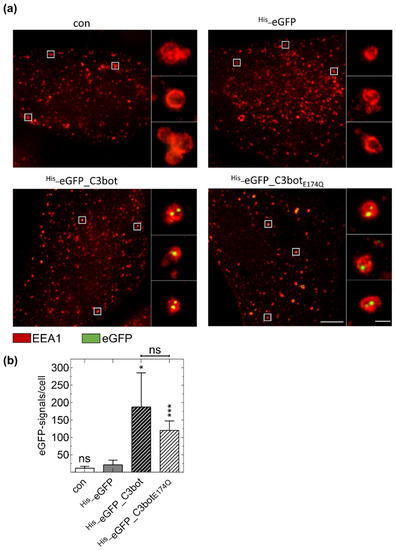
Clostridial C3 Toxins Enter and Intoxicate HumanDendritic Cells
Maximilian Fellermann, Christina Huchler, Lea Fechter, Tobias Kolb, Fanny Wondany, Daniel Mayer, Jens Michaelis, Steffen Stenger, Kevin Mellert, Peter Möller, Thomas F. E. Barth, Stephan Fischer and Holger Barth
C3 protein toxins produced byClostridium (C.) botulinumandC. limosumare mono-ADP-ribosyltransferases, which specifically modify the GTPases Rho A/B/C in the cytosol of monocytic cells,thereby inhibiting Rho-mediated signal transduction in monocytes, macrophages, and osteoclasts.C3 toxins are selectively taken up into the cytosol of monocytic cells by endocytosis and translocatefrom acidic endosomes into the cytosol. The C3-catalyzed ADP-ribosylation of Rho proteins inhibitsessential functions of these immune cells, such as migration and phagocytosis. Here, we demonstratethat C3 toxins enter and intoxicate dendritic cells in a time- and concentration-dependent manner.Both immature and mature human dendritic cells efficiently internalize C3 exoenzymes. These findingscould also be extended to the chimeric fusion toxin C2IN-C3lim. Moreover, stimulated emissiondepletion (STED) microscopy revealed the localization of the internalized C3 protein in endosomesand emphasized its potential use as a carrier to deliver foreign proteins into dendritic cells. In contrast,the enzyme C2I from the binaryC. botulinumC2 toxin was not taken up into dendritic cells, indicatingthe specific uptake of C3 toxins. Taken together, we identified human dendritic cells as novel target cellsfor clostridial C3 toxins and demonstrated the specific uptake of these toxins via endosomal vesicles.
Toxins
https://www.mdpi.com/2072-6651/12/9/563

Determination of the 3D Magnetic Field Vector Orientation with NV Centers in Diamond
Timo Weggler, Christian Ganslmayer, Florian Frank, Tobias Eilert, Fedor Jelezko, Jens Michaelis
Absolute knowledge about the magnetic field orientation plays a crucial role in single spin-based quantum magnetometry and the application toward spin-based quantum computation. In this paper, we reconstruct the 3D orientation of an arbitrary static magnetic field with individual nitrogen vacancy (NV) centers in diamond. We determine the polar and the azimuthal angle of the magnetic field orientation relative to the diamond lattice. Therefore, we use information from the photoluminescence anisotropy of the NV, together with a simple pulsed Optically Detected Magnetic Resonance (ODMR) experiment. Our nanoscopic magnetic field determination is generally applicable and does not rely on special prerequisites such as strongly coupled nuclear spins or particular controllable fields. Hence, our presented results open up new paths for precise NMR reconstructions and the modulation of the electron-electron spin interaction in EPR measurements by specifically tailored magnetic fields.
Nano Lett.
https://pubs.acs.org/doi/abs/10.1021/acs.nanolett.9b04725
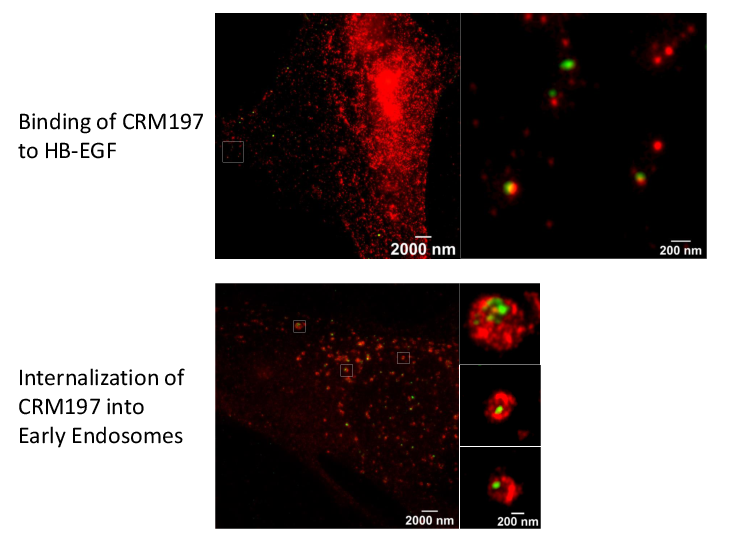
Super-resolution microscopy unveils transmembrane domain-mediated internalization of cross-reacting material 197 into diphtheria toxin-resistant mouse J774A.1 cells and primary rat fibroblasts in vitro
Maximilian Fellermann, Fanny Wondany, Stefan Carle, Julia Nemeth, Tanmay Sadhanasatish, Manfred Frick, Holger Barth, Jens Michaelis
Diphtheria toxin (DT) efficiently inhibits protein synthesis in human cells, resulting in severe disease diphtheria. The sensitivity towards DT varies between mammalian species. Mice and rats are resistant to DT. However, the reason underlying this insensitivity is controversially discussed and not well understood. Therefore, we investigated the steps of DT uptake, i.e. receptor binding and internalization into mouse J774A.1 macrophages and primary rat fibroblasts. We exploited the non-toxic DT-mutant cross-reacting material 197 (CRM197) and three additional receptor binding-deficient mutants (250 nM each) to investigate binding to cell surface and internalization into murine cells via flow cytometry and stimulated emission depletion (STED) super-resolution optical microscopy. Dual-color STED imaging unveiled CRM197 interacting with the murine precursor of the heparin-binding epidermal growth factor-like growth factor (HB-EGF). Moreover, we identified CRM197’s transmembrane domain as an additional HB-EGF binding site, which is also involved in the receptor-mediated internalization into murine cells. However, we do not find evidence for translocation of the catalytically active subunit (DTA) into the cytosol when 250 nM DT were applied. In conclusion, we provide evidence that the resistance of murine cells to DT is caused by an insufficiency of DTA to escape from endosomes and reach the cytosol. Possibly, a higher affinity interaction of DT and the HB-EGF is required for translocation, which highlights the role of the receptor in the endosomes during the translocation step. We extend the current knowledge about cellular uptake of the medically relevant DT and CRM197.
Archives of Toxicology
https://doi.org/10.1007/s00204-020-02731-4

Myosin VI moves on nuclear actin filaments and supports long-range chromatin rearrangements
Andreas Große-Berkenbusch, Johannes Hettich, Timo Kuhn, Natalia Fili, Alexander W. Cook, Yukti Hari-Gupta, Anja Palmer, Lisa Streit, Peter J.I. Ellis, Christopher P. Toseland and J. Christof M. Gebhardt
Nuclear myosin VI (MVI) enhances RNA polymerase II – dependent transcription, but the molecular mechanism is unclear. We used live cell single molecule tracking to follow individual MVI molecules inside the nucleus and observed micrometer-long motion of the motor. Besides static chromatin interactions lasting for tens of seconds, ATPase-dependent directed motion occurred with a velocity of 2 µm/s. The movement was frequently interrupted by short periods of slow restricted diffusion and increased in frequency upon stimulation of transcription. Mutagenesis and perturbation experiments demonstrated that nuclear MVI motion is independent of dimerization and occurs on nuclear actin filaments, which we also observed by two-color imaging. Using chromosome paint to quantify distances between chromosomes, we found that MVI is required for transcription-dependent long-range chromatin rearrangements. Our measurements reveal a transcription-coupled function of MVI in the nucleus, where it actively undergoes directed movement along nuclear actin filaments. Motion is potentially mediated by cooperating monomeric motors and might assist in enhancing transcription by supporting long-range chromatin rearrangements.
bioRxiv
https://doi.org/10.1101/2020.04.03.023614
Use of Super-Resolution Optical Microscopy To Reveal Direct Virus Binding at Hybrid Core−Shell Matrixes
Manuela Gast, Fanny Wondany, Bastian Raabe, Jens Michaelis, Harald Sobek, and Boris Mizaikoff
Polymer particles with antibody-like affinity, i.e., molecularly imprinted polymers, offer an ideal platform for biopharmaceutical virus purification. In recent years, attempts combining molecular imprinting technology with a variety of visualization and detection techniques have been reported for directly confirming the localized presence of the template. Direct target visualization is crucial for the characterization of molecularly imprinted polymers, especially if biological templates such as viruses are used. In the present study, for the first time the viral binding behavior at virus-imprinted polymers (VIPs) via stimulated emission depletion (STED) microscopy is shown by imaging individual, fluorescently labeled virus particles. STED microscopy achieves among various other super-resolution techniques the best temporal resolution at high spatial resolution. An innovative virus purification material selective for human adenovirus type 5 (AdV5) offered highly purified virus for the subsequent fluorescent labeling procedure, thus enabling STED imaging. Excellent binding affinities (150-fold higher versus control particles) and high selectivity toward the target virus (AdV5) were observed at those VIPs, even in competitive binding experiments with minute virus of mice using dual-label STED microscopy.
Anal. Chem. 2020
https://doi.org/10.1021/acs.analchem.9b04328

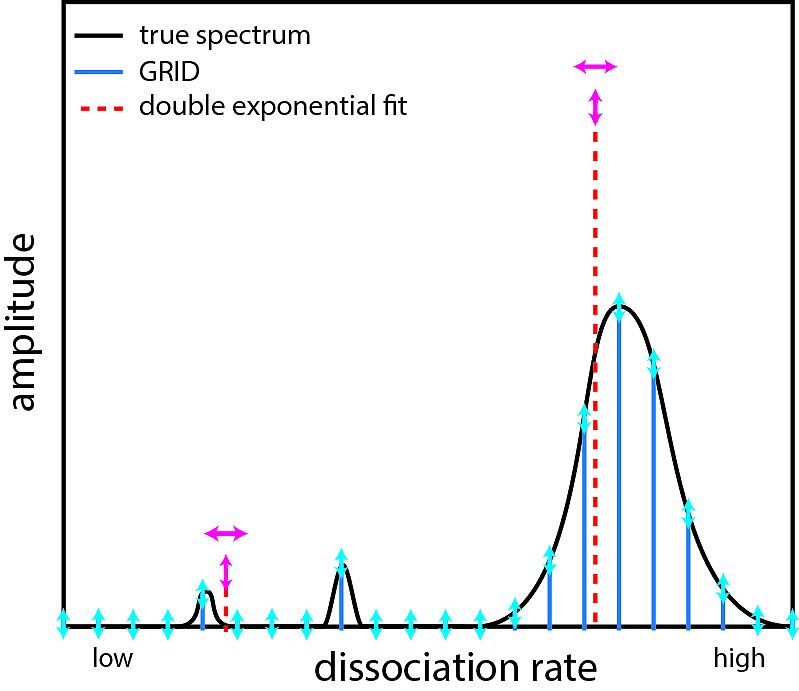
Inferring quantity and qualities of superimposed reaction rates from single molecule survival time distributions
Matthias Reisser, Johannes Hettich, Timo Kuhn, Achim P. Popp, Andreas Große-Berkenbusch & J. Christof M. Gebhardt
Actions of molecular species, for example binding of transcription factors to chromatin, may comprise several superimposed reaction pathways. The number and the rate constants of such superimposed reactions can in principle be resolved by inverse Laplace transformation of the corresponding distribution of reaction lifetimes. However, current approaches to solve this transformation are challenged by photobleaching-prone fluorescence measurements of lifetime distributions. Here, we present a genuine rate identification method (GRID), which infers the quantity, rates and amplitudes of dissociation processes from fluorescence lifetime distributions using a dense grid of possible decay rates. In contrast to common multi-exponential analysis of lifetime distributions, GRID is able to distinguish between broad and narrow clusters of decay rates. We validate GRID by simulations and apply it to CDX2-chromatin interactions measured by live cell single molecule fluorescence microscopy. GRID reveals well-separated narrow decay rate clusters of CDX2, in part overlooked by multi-exponential analysis. We discuss the amplitudes of the decay rate spectrum in terms of frequency of observed events and occupation probability of reaction states. We further demonstrate that a narrow decay rate cluster is compatible with a common model of TF sliding on DNA.
Scientific Reports
https://www.nature.com/articles/s41598-020-58634-y