Single molecule biophysics in living cells
Current research:
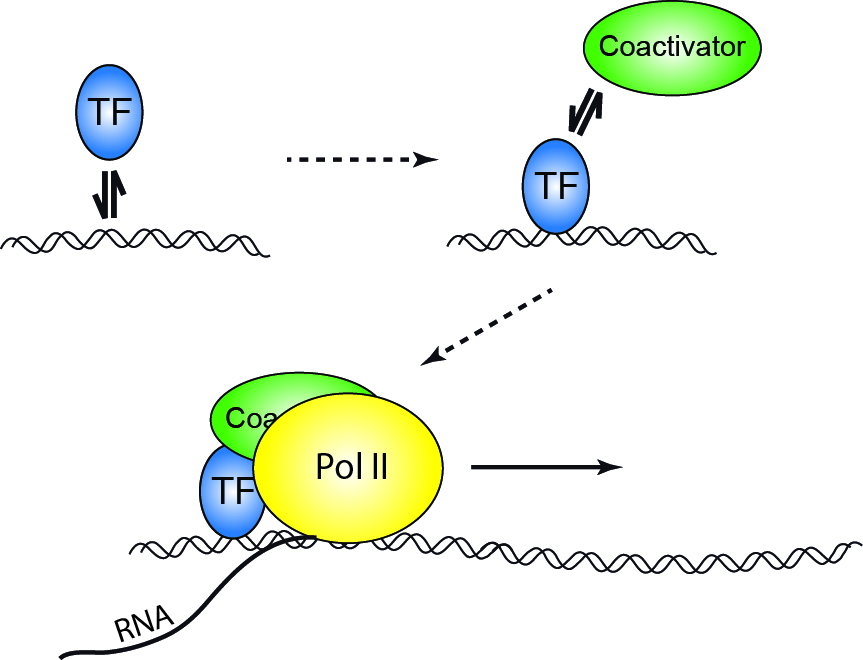
Binding of a transcription factor to a regulatory sequence on DNA is the first step in gene expression, accompanied by binding of different cofactors and enzymes, assembly of the transcription complex and final read-out of the genetic information stored in genomic DNA. Interestingly, the interactions between initiating factors and DNA are short-lived and stochastic. This raises questions about the temporal coordination between binding events as well as the coupling between transcription factor binding and the onset of transcription:
How does the cell carry forward the information of a first binding event to eventual expression of a gene?
How do cofactors influence the temporal sequence?
Does binding of a transcription factor initiate an independently running process, or is continuous transcription accompanied by repeated transcription factor binding?
We approach these intriguing questions by following individual biomolecules involved in gene expression in their natural environment of a living cell and study their temporal interplay with other components of transcription using multi color single molecule fluorescence microscopy.
Published projects:
→ Chromatin Architecture
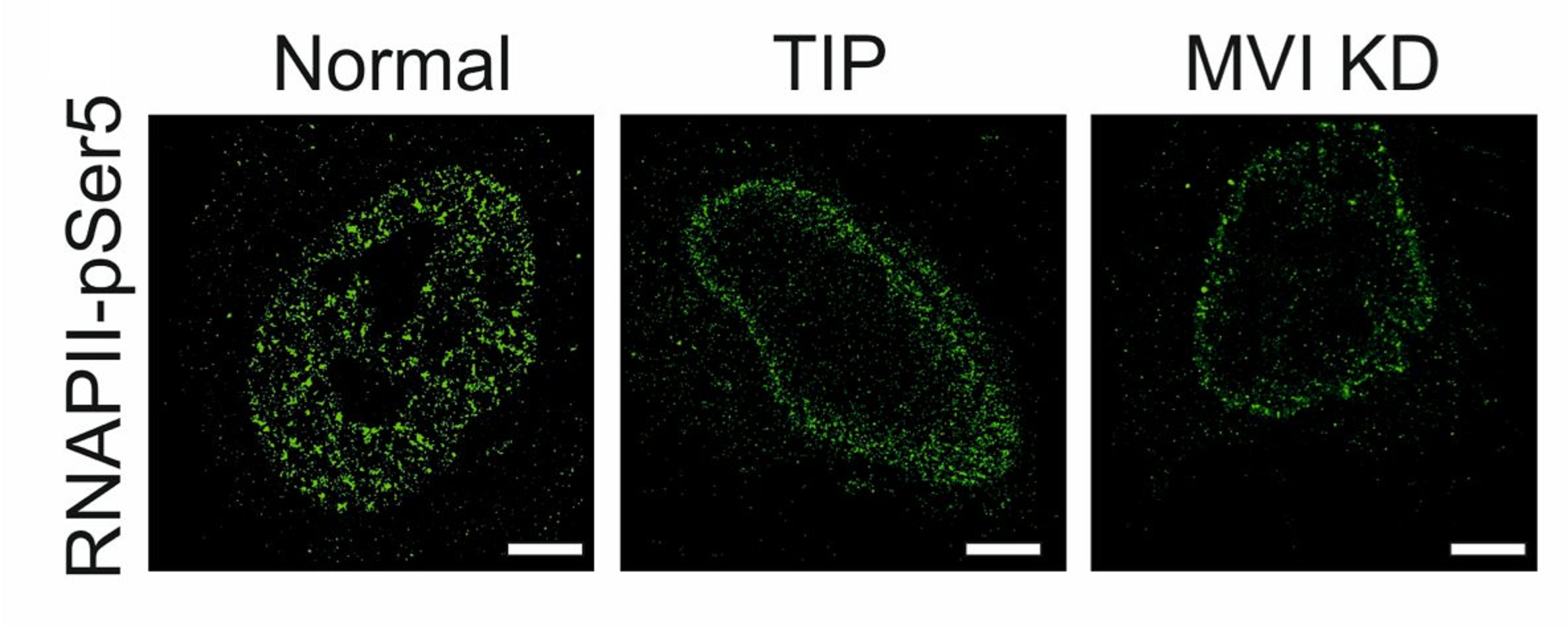
Myosin VI regulates the spatial organisation of mammalian transcription initiatio
During transcription, RNA Polymerase II (RNAPII) is spatially organised within the nucleus into clusters that correlate with transcription activity. While this is a hallmark of genome regulation in mammalian cells, the mechanisms concerning the assembly, organisation and stability remain unknown. Here, we have used combination of single molecule imaging and genomic approaches to explore the role of nuclear myosin VI (MVI) in the nanoscale organisation of RNAPII. We reveal that MVI in the nucleus acts as the molecular anchor that holds RNAPII in high density clusters. Perturbation of MVI leads to the disruption of RNAPII localisation, chromatin organisation and subsequently a decrease in gene expression. Overall, we uncover the fundamental role of MVI in the spatial regulation of gene expression.
https://doi.org/10.1038/s41467-022-28962-w
Myosin VI moves on nuclear actin filaments and supports long-range chromatin rearrangements
Nuclear myosin VI (MVI) enhances RNA polymerase II – dependent transcription, but the molecular mechanism is unclear. We used live cell single molecule tracking to follow individual MVI molecules inside the nucleus and observed micrometer-long motion of the motor. Besides static chromatin interactions lasting for tens of seconds, ATPase-dependent directed motion occurred with a velocity of 2 µm/s. The movement was frequently interrupted by short periods of slow restricted diffusion and increased in frequency upon stimulation of transcription. Mutagenesis and perturbation experiments demonstrated that nuclear MVI motion is independent of dimerization and occurs on nuclear actin filaments, which we also observed by two-color imaging. Using chromosome paint to quantify distances between chromosomes, we found that MVI is required for transcription-dependent long-range chromatin rearrangements. Our measurements reveal a transcription-coupled function of MVI in the nucleus, where it actively undergoes directed movement along nuclear actin filaments. Motion is potentially mediated by cooperating monomeric motors and might assist in enhancing transcription by supporting long-range chromatin rearrangements.

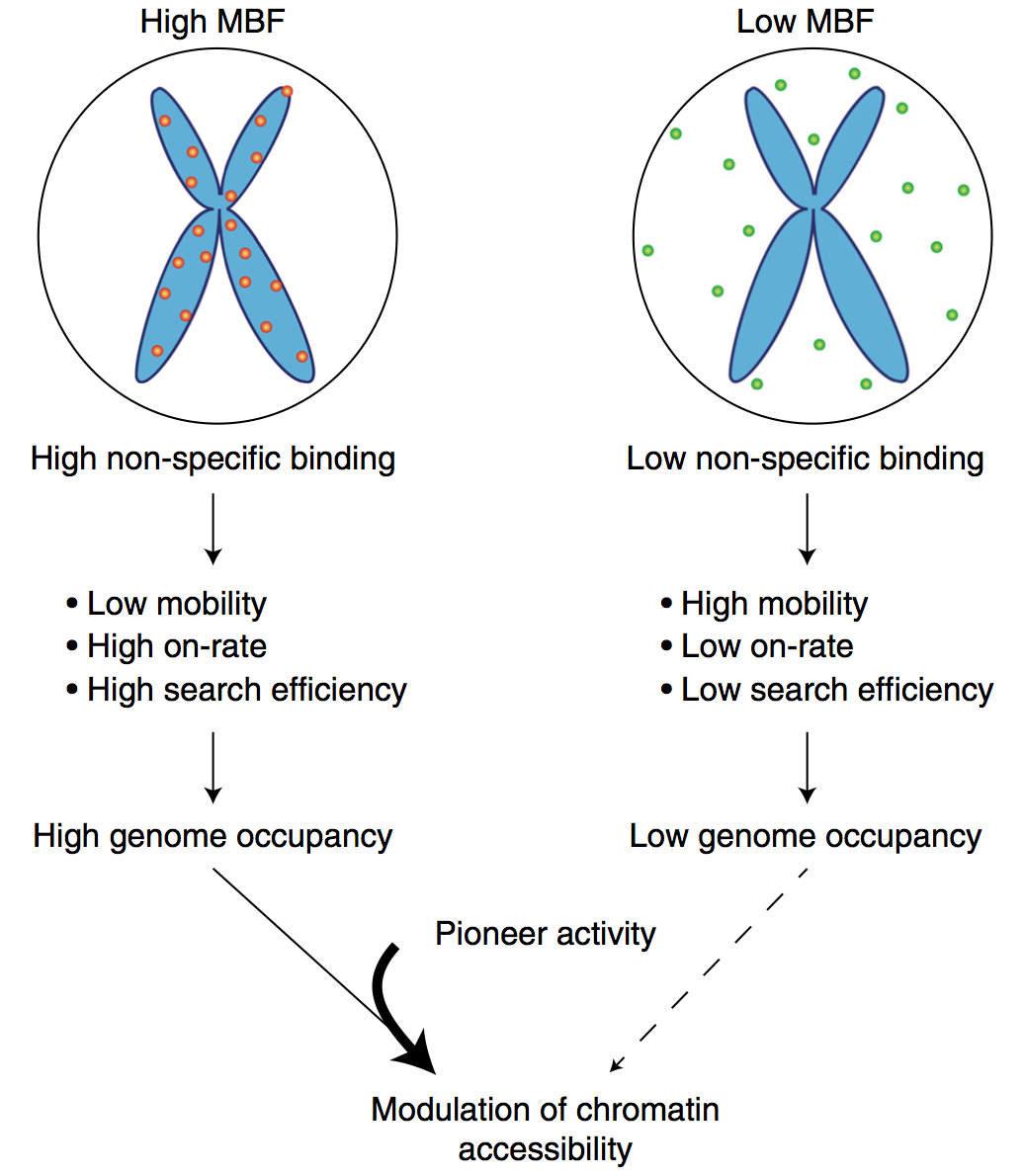
Mitotic chromosome binding predicts transcription factor properties in interphase
Mammalian transcription factors (TFs) differ broadly in their nuclear mobility and sequence- specific/non-specific DNA binding. How these properties affect their ability to occupy specific genomic sites and modify the epigenetic landscape is unclear. The association of TFs with mitotic chromosomes observed by fluorescence microscopy is largely mediated by non-specific DNA interactions and differs broadly between TFs. Here we combine quantitative measure- ments of mitotic chromosome binding (MCB) of 501 TFs, TF mobility measurements by fluorescence recovery after photobleaching, single molecule imaging of DNA binding, and mapping of TF binding and chromatin accessibility. TFs associating to mitotic chromosomes are enriched in DNA-rich compartments in interphase and display slower mobility in interphase and mitosis. Remarkably, MCB correlates with relative TF on-rates and genome-wide specific site occupancy, but not with TF residence times. This suggests that non-specific DNA binding properties of TFs regulate their search efficiency and occupancy of specific genomic sites.
https://www.biorxiv.org/content/early/2018/08/31/404723
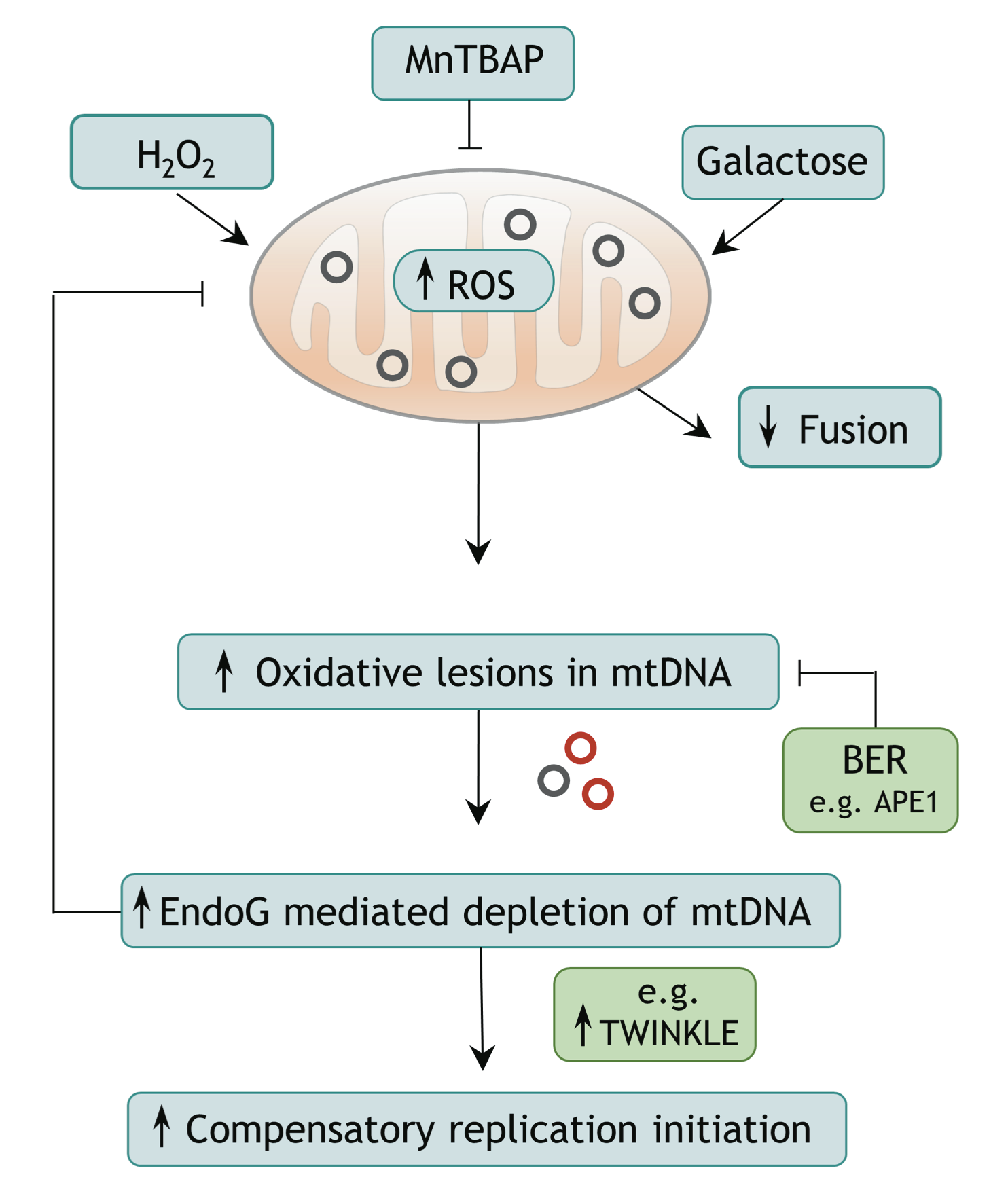
Endonuclease G promotes mitochondrial genome cleavage and replication
Endonuclease G (EndoG) is a nuclear-encoded endonuclease, mostly localised in mitochondria. In the nucleus EndoG participates in site-specific cleavage during replication stress and genome-wide DNA degradation during apoptosis. However, the impact of EndoG on mitochondrial DNA (mtDNA) metabolism is poorly understood. Here, we investigated whether EndoG is involved in the regulation of mtDNA replication and removal of aberrant copies. We applied the single-cell mitochondrial Transcription and Replication Imaging Protocol (mTRIP) and PCR-based strategies on human cells after knockdown/knockout and re-expression of EndoG. Our analysis revealed that EndoG stimulates both mtDNA replication initiation and mtDNA depletion, the two events being interlinked and dependent on EndoG’s nuclease activity. Stimulation of mtDNA replication by EndoG was independent of 7S DNA processing at the replication origin. Importantly, both mtDNA-directed activities of EndoG were promoted by oxidative stress. Inhibition of base excision repair (BER) that repairs oxidative stress- induced DNA damage unveiled a pronounced effect of EndoG on mtDNA removal, reminiscent of recently discovered links between EndoG and BER in the nucleus. Altogether with the downstream effects on mitochondrial transcription, protein expression, redox status and morphology, this study demonstrates that removal of damaged mtDNA by EndoG and compensatory replication play a critical role in mitochondria homeostasis.
doi.org/10.18632/oncotarget.24822
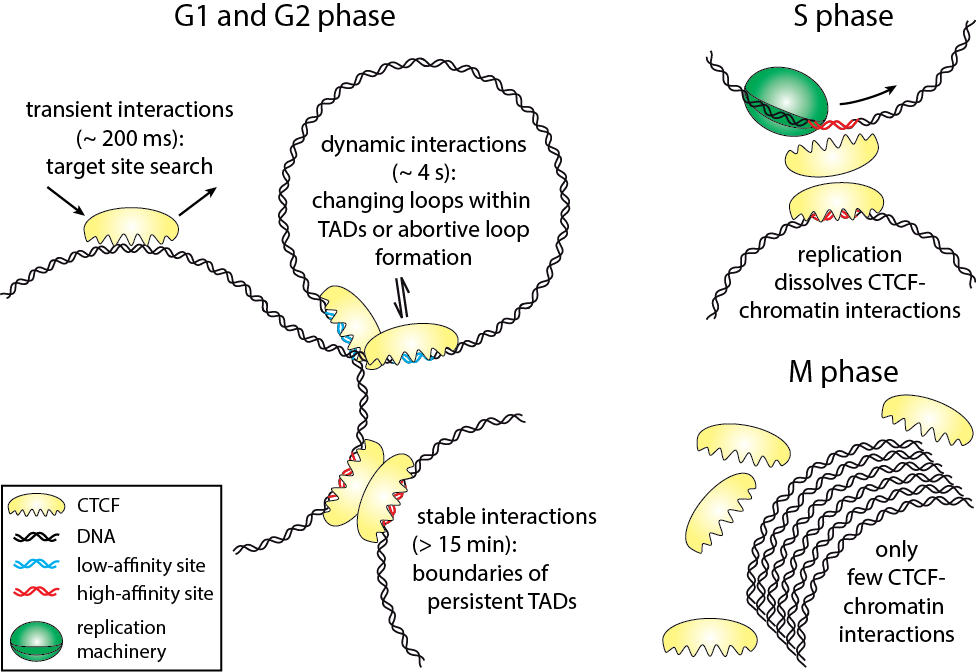
Cell-cycle-dependent interactions between CTCF and Chromatin
The three-dimensional arrangement of chromatin encodes regulatory traits important for nuclear processes such as transcription and replication. Chromatin topology is in part mediated by the architectural protein CCCTC-binding factor (CTCF) that binds to the boundaries of topologically associating domains. Whereas sites of CTCF interactions are well characterized, little is known on how long CTCF binds to chromatin and how binding evolves during the cell cycle. We monitored CTCF-chromatin interactions by live cell single molecule tracking in different phases of the cell cycle. In G1-, S-, and G2-phases, a majority of CTCF molecules was bound transiently (∼0.2 s) to chromatin, whereas minor fractions were bound dynamically (∼4 s) or stably (>15 min). During mitosis, CTCF was mostly excluded from chromatin. Our data suggest that CTCF scans DNA in search for two different subsets of specific target sites and provide information on the timescales over which topologically associating domains might be restructured. During S-phase, dynamic and stable interactions decreased considerably compared to G1-phase, but were resumed in G2-phase, indicating that specific interactions need to be dissolved for replication to proceed.
Biophysical Journal 112, 1-5, May 23, 2017
→ Transcription Regulation
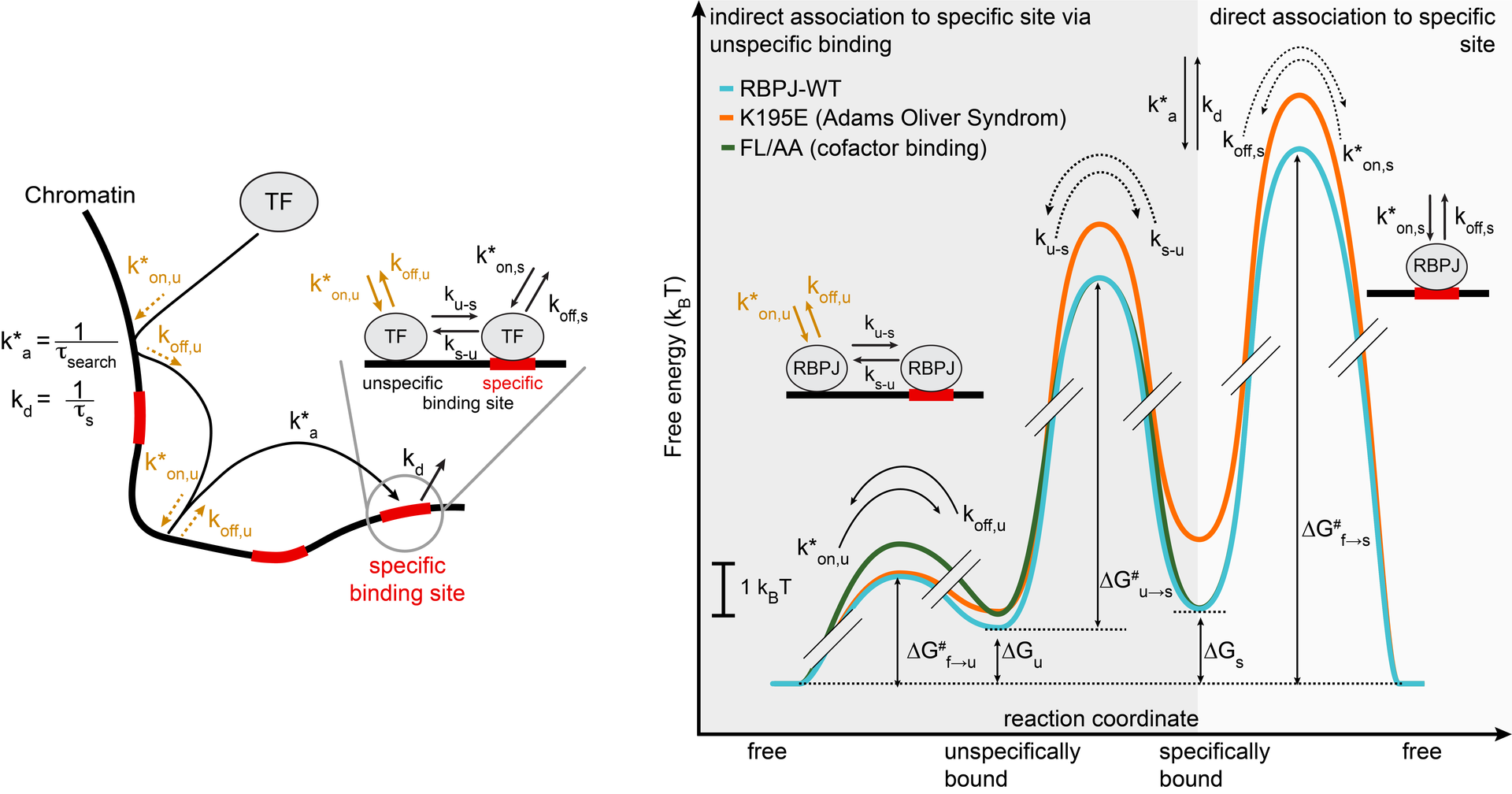
In vivo binding free energy landscape reveals kinetic control of transcription factor function
Transcription factors such as RBPJ in Notch signal transduction bind to specific DNA sequences and initiate either repression or activation of genes. Which sites they select and how often and long they bind affects the efficiency of gene regulation. To resolve the underpinnings of RBPJ-DNA binding, we determined the in vivo binding free energy landscape of RBPJ using live-cell single-molecule tracking and genome-wide chromatin immunoprecipitation. Importantly, DNA binding of RBPJ was thermodynamically unstable in vivo and instead governed by the binding kinetics: Cofactors contributed to target site specificity by tuning both association and dissociation of unspecific binding, while mutation K195E underlying Adams-Oliver-Syndrome destabilized specific DNA binding by mainly altering the association rate. We showed thermodynamic instability in vivo also for other transcription factors, indicating that kinetic rather than thermodynamic control of DNA binding might be a general feature of transcription factors in vivo.
www.biorxiv.org/content/10.1101/2023.12.19.572376v1
Transcriptional reprogramming by mutated IRF4 in lymphoma
Disease-causing mutations in genes encoding transcription factors (TFs) can affect TF interactions with their cognate DNA-binding motifs. Whether and how TF mutations impact upon the binding to TF composite elements (CE) and the interaction with other TFs is unclear. Here, we report a distinct mechanism of TF alteration in human lymphomas with perturbed B cell identity, in particular classic Hodgkin lymphoma. It is caused by a recurrent somatic missense mutation c.295 T > C (p.Cys99Arg; p.C99R) targeting the center of the DNA-binding domain of Interferon Regulatory Factor 4 (IRF4), a key TF in immune cells. IRF4-C99R fundamentally alters IRF4 DNA-binding, with loss-of-binding to canonical IRF motifs and neomorphic gain-of-binding to canonical and non-canonical IRF CEs. IRF4-C99R thoroughly modifies IRF4 function by blocking IRF4-dependent plasma cell induction, and up-regulates disease-specific genes in a non-canonical Activator Protein-1 (AP-1)-IRF-CE (AICE)-dependent manner. Our data explain how a single mutation causes a complex switch of TF specificity and gene regulation and open the perspective to specifically block the neomorphic DNA-binding activities of a mutant TF.
https://doi.org/10.1038/s41467-023-41954-8
A multimorphic mutation in IRF4 causes human autosomal dominant combined immunodeficiency
Interferon regulatory factor 4 (IRF4) is a transcription factor (TF) and key regulator of immune cell development and function. We report a recurrent heterozygous mutation in IRF4, p.T95R, causing an autosomal dominant combined immunodeficiency (CID) in seven patients from six unrelated families. The patients exhibited profound susceptibility to opportunistic infections, notably Pneumocystis jirovecii, and presented with agammaglobulinemia. Patients’ B cells showed impaired maturation, decreased immunoglobulin isotype switching, and defective plasma cell differentiation, whereas their T cells contained reduced TH17 and TFH populations and exhibited decreased cytokine production. A knock-in mouse model of heterozygous T95R showed a severe defect in antibody production both at the steady state and after immunization with different types of antigens, consistent with the CID observed in these patients. The IRF4T95R variant maps to the TF’s DNA binding domain, alters its canonical DNA binding specificities, and results in a simultaneous multimorphic combination of loss, gain, and new functions for IRF4. IRF4T95R behaved as a gain-of-function hypermorph by binding to DNA with higher affinity than IRF4WT. Despite this increased affinity for DNA, the transcriptional activity on IRF4 canonical genes was reduced, showcasing a hypomorphic activity of IRF4T95R. Simultaneously, IRF4T95R functions as a neomorph by binding to noncanonical DNA sites to alter the gene expression profile, including the transcription of genes exclusively induced by IRF4T95R but not by IRF4WT. This previously undescribed multimorphic IRF4 pathophysiology disrupts normal lymphocyte biology, causing human disease.
https://doi.org/10.1126/sciimmunol.ade7953
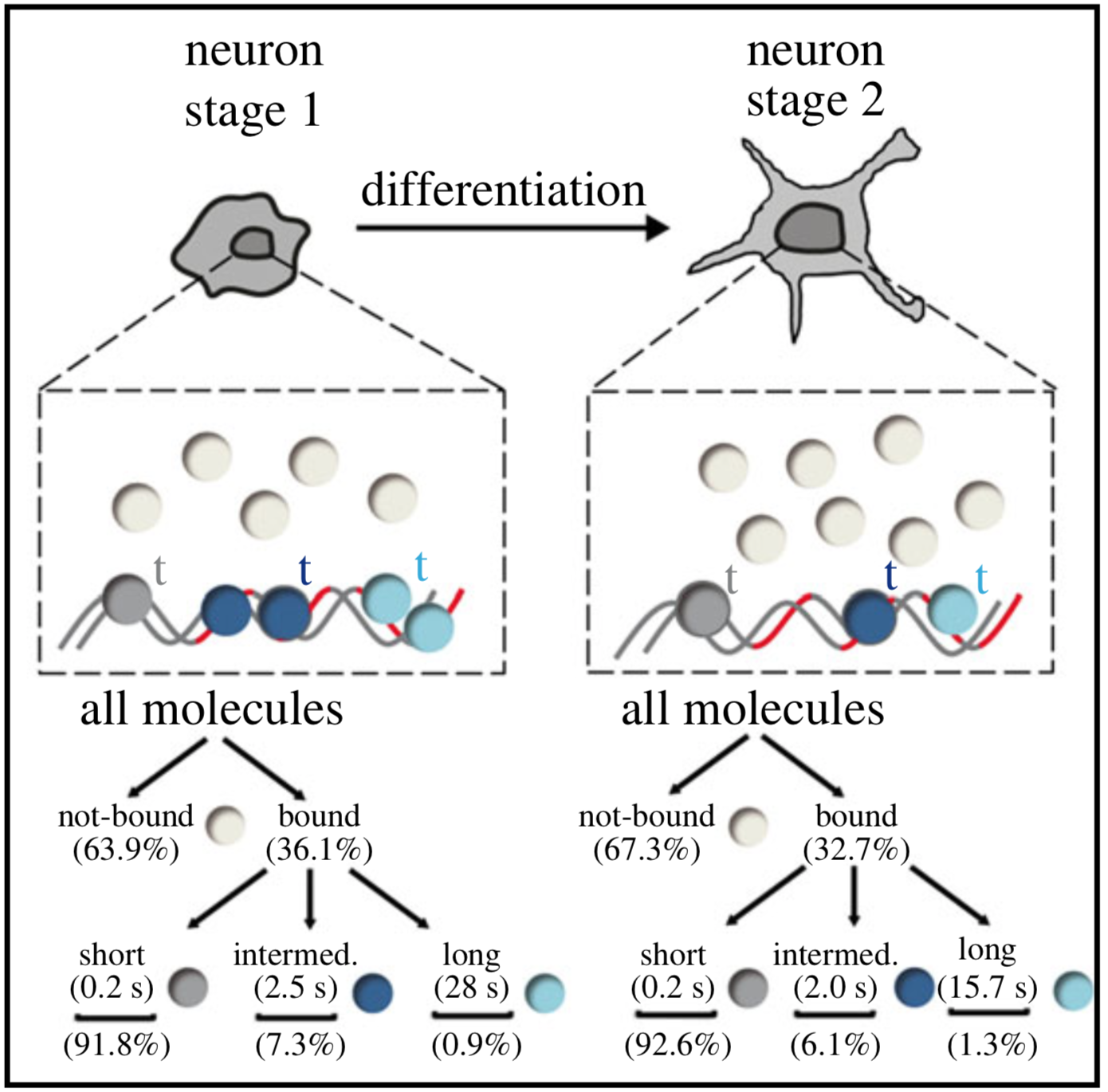
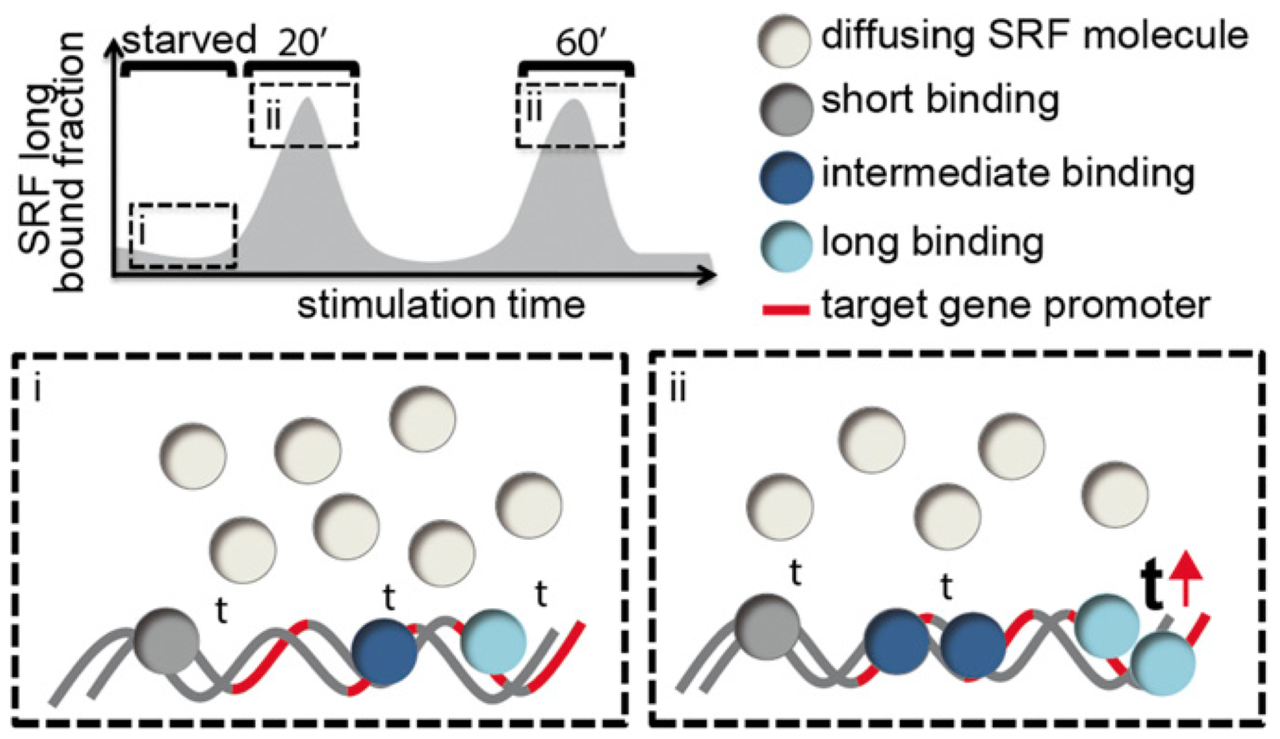
Single-molecule tracking (SMT) and localization of SRF and MRTF transcription factors during neuronal stimulation and differentiation
In cells, proteins encoded by the same gene do not all behave uniformly but engage in functional subpopulations induced by spatial or temporal segregation. While conventional microscopy has limitations in revealing such spatial and temporal diversity, single-molecule tracking (SMT) microscopy circumvented this problem and allows for high-resolution imaging and quantification of dynamic single-molecule properties. Particularly in the nucleus, SMT has identified specific DNA residence times of transcription factors (TFs), DNA-bound TF fractions and positions of transcriptional hot-spots upon cell stimulation. By contrast to cell stimulation, SMT has not been employed to follow dynamic TF changes along stages of cell differentiation. Herein, we analysed the serum response factor (SRF), a TF involved in the differentiation of many cell types to study nuclear single-molecule dynamics in neuronal differentiation. Our data in living mouse hippocampal neurons show dynamic changes in SRF DNA residence time and SRF DNA-bound fraction between the stages of adhesion, neurite growth and neurite differentiation in axon and dendrites. Using TALM (tracking and localization microscopy), we identified nuclear positions of SRF clusters and observed changes in their numbers and size during differentiation. Furthermore, we show that the SRF cofactor MRTF-A (myocardin-related TF or MKL1) responds to cell activation by enhancing the long-bound DNA fraction. Finally, a first SMT colocalization study of two proteins was performed in living cells showing enhanced SRF/MRTF-A colocalization upon stimulation. In summary, SMT revealed modulation of dynamic TF properties during cell stimulation and differentiation.
https://doi.org/10.1098/rsob.210383
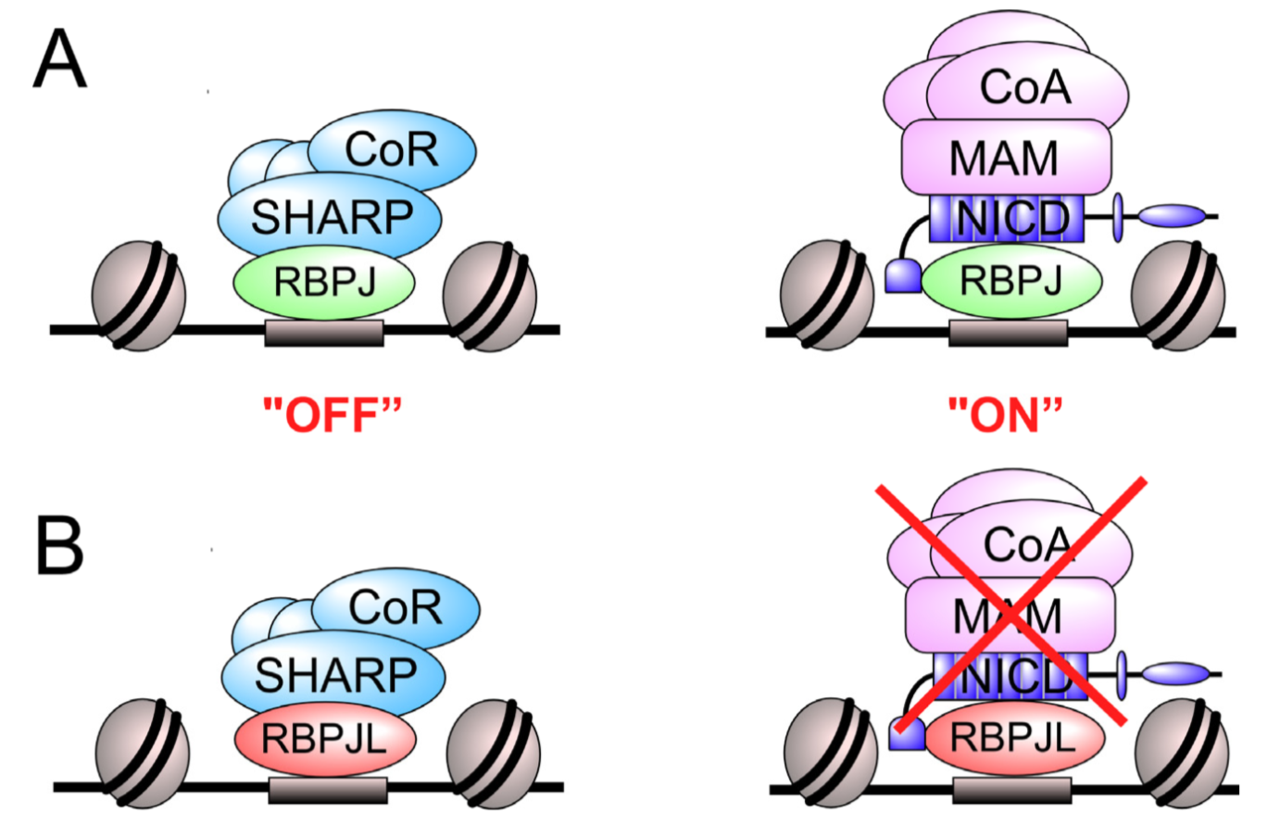
Transcription Factor RBPJL Is Able to Repress Notch Target Gene Expression but Is Non-Responsive to Notch Activation
The Notch signaling pathway is an evolutionary conserved signal transduction cascade present in almost all tissues and is required for embryonic and postnatal development, as well as for stem cell maintenance, but it is also implicated in tumorigenesis including pancreatic cancer and leukemia. The transcription factor RBPJ forms a coactivator complex in the presence of a Notch signal, whereas it represses Notch target genes in the absence of a Notch stimulus. In the pancreas, a specific paralog of RBPJ, called RBPJL, is expressed and found as part of the heterotrimeric PTF1-complex. However, the function of RBPJL in Notch signaling remains elusive. Using molecular modeling, biochemical and functional assays, as well as single-molecule time-lapse imaging, we show that RBPJL and RBPJ, despite limited sequence homology, possess a high degree of structural similarity. RBPJL is specifically expressed in the exocrine pancreas, whereas it is mostly undetectable in pancreatic tumour cell lines. Importantly, RBPJL is not able to interact with Notch−1 to −4 and it does not support Notch-mediated transactivation. However, RBPJL can bind to canonical RBPJ DNA elements and shows migration dynamics comparable to that of RBPJ in the nuclei of living cells. Importantly, RBPJL is able to interact with SHARP/SPEN, the central corepressor of the Notch pathway. In line with this, RBPJL is able to fully reconstitute transcriptional repression at Notch target genes in cells lacking RBPJ. Together, RBPJL can act as an antagonist of RBPJ, which renders cells unresponsive to the activation of Notch.
https://www.mdpi.com/2072-6694/13/19/5027
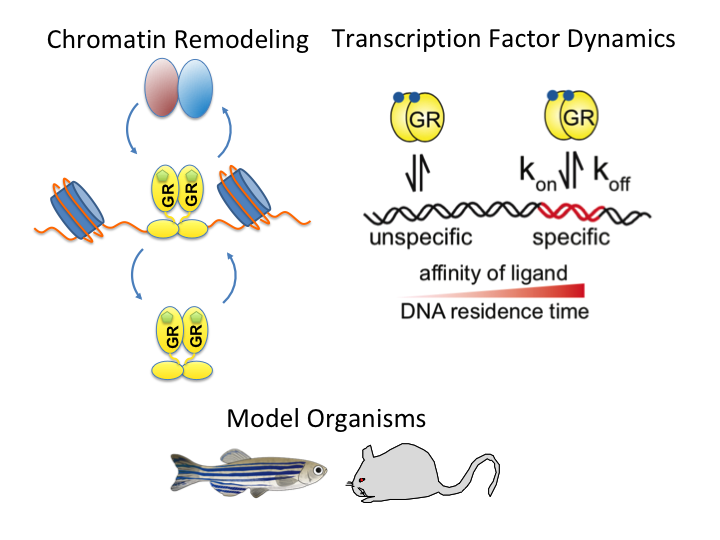
A guide to changing paradigms of glucocorticoid receptor function—a model system for genome regulation and physiology
The glucocorticoid receptor (GR) is a bona fide ligand-regulated transcription factor. Cloned in the 80s, the GR has become one of the best-studied and clinically most relevant members of the nuclear receptor superfamily. Cooperative activity of GR with other transcription factors and a plethora of coregulators contribute to the tissue- and context-specific response toward the endogenous and pharmacological glucocorticoids (GCs). Furthermore, nontranscriptional activities in the cytoplasm are emerging as an additional function of GR. Over the past 40 years, the concepts of GR mechanisms of action had been constantly changing. Different methodologies in the pregenomic and genomic era of molecular biological research and recent cutting-edge technology in single-cell and single-molecule analysis are steadily evolving the views, how the GR in particular and transcriptional regulation in general act in physiological and pathological processes. In addition to the development of technologies for GR analysis, the use of model organisms provides insights how the GR in vivo executes GC action in tissue homeostasis, inflammation, and energy metabolism. The model organisms, namely the mouse, but also rats, zebrafish, and recently fruit flies carrying mutations of the GR became a major driving force to analyze the molecular function of GR in disease models. This guide provides an overview of the exciting research and paradigm shifts in the GR field from past to present with a focus on GR transcription factor networks, GR DNA-binding and single-cell analysis, and model systems.
https://doi.org/10.1111/febs.16100
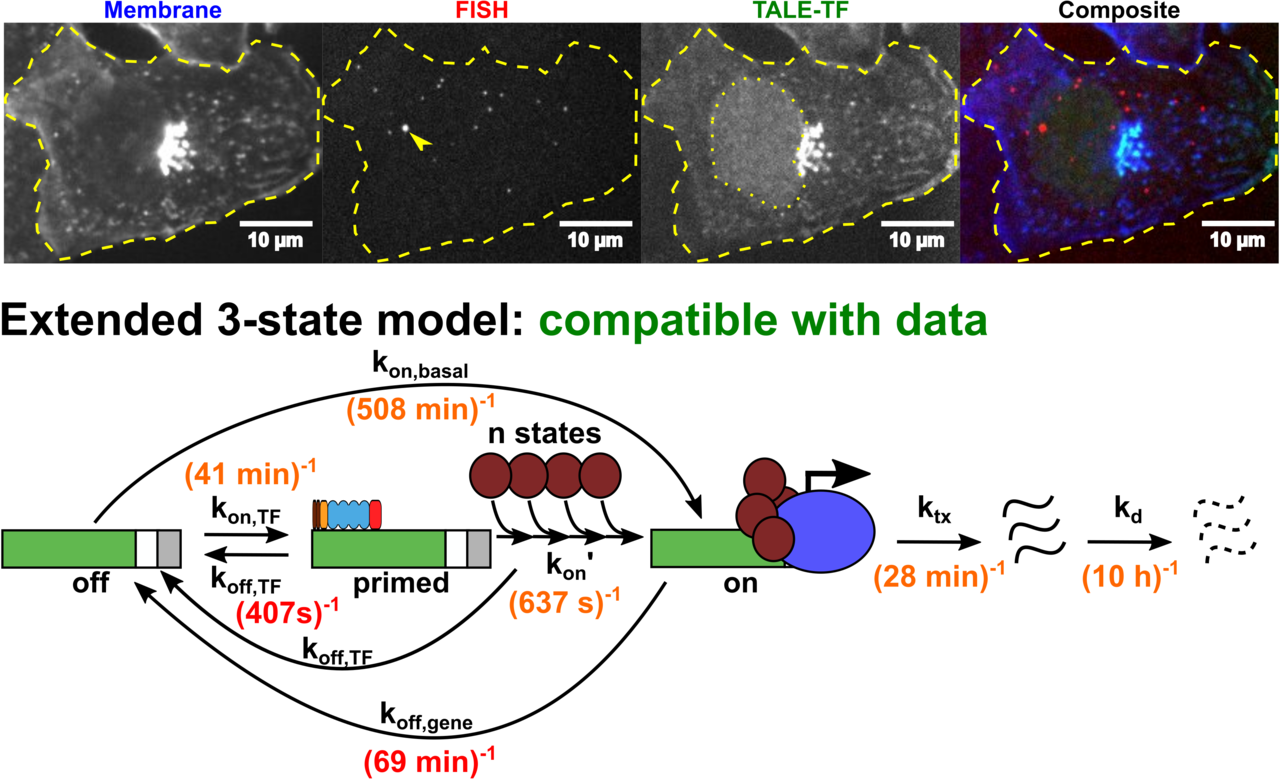
Altering transcription factor binding reveals comprehensive transcriptional kinetics of a basic gene
Transcription is a vital process activated by transcription factor (TF) binding. The active gene releases a burst of transcripts before turning inactive again. While the basic course of transcription is well understood, it is unclear how binding of a TF affects the frequency, duration and size of a transcriptional burst. We systematically varied the residence time and concentration of a synthetic TF and characterized the transcription of a synthetic reporter gene by combining single molecule imaging, single molecule RNA-FISH, live transcript visualisation and analysis with a novel algorithm, Burst Inference from mRNA Distributions (BIRD). For this well-defined system, we found that TF binding solely affected burst frequency and variations in TF residence time had a stronger influence than variations in concentration. This enabled us to device a model of gene transcription, in which TF binding triggers multiple successive steps before the gene transits to the active state and actual mRNA synthesis is decoupled from TF presence. We quantified all transition times of the TF and the gene, including the TF search time and the delay between TF binding and the onset of transcription. Our quantitative measurements and analysis revealed detailed kinetic insight, which may serve as basis for a bottom-up understanding of gene regulation.
https://doi.org/10.1093/nar/gkab443
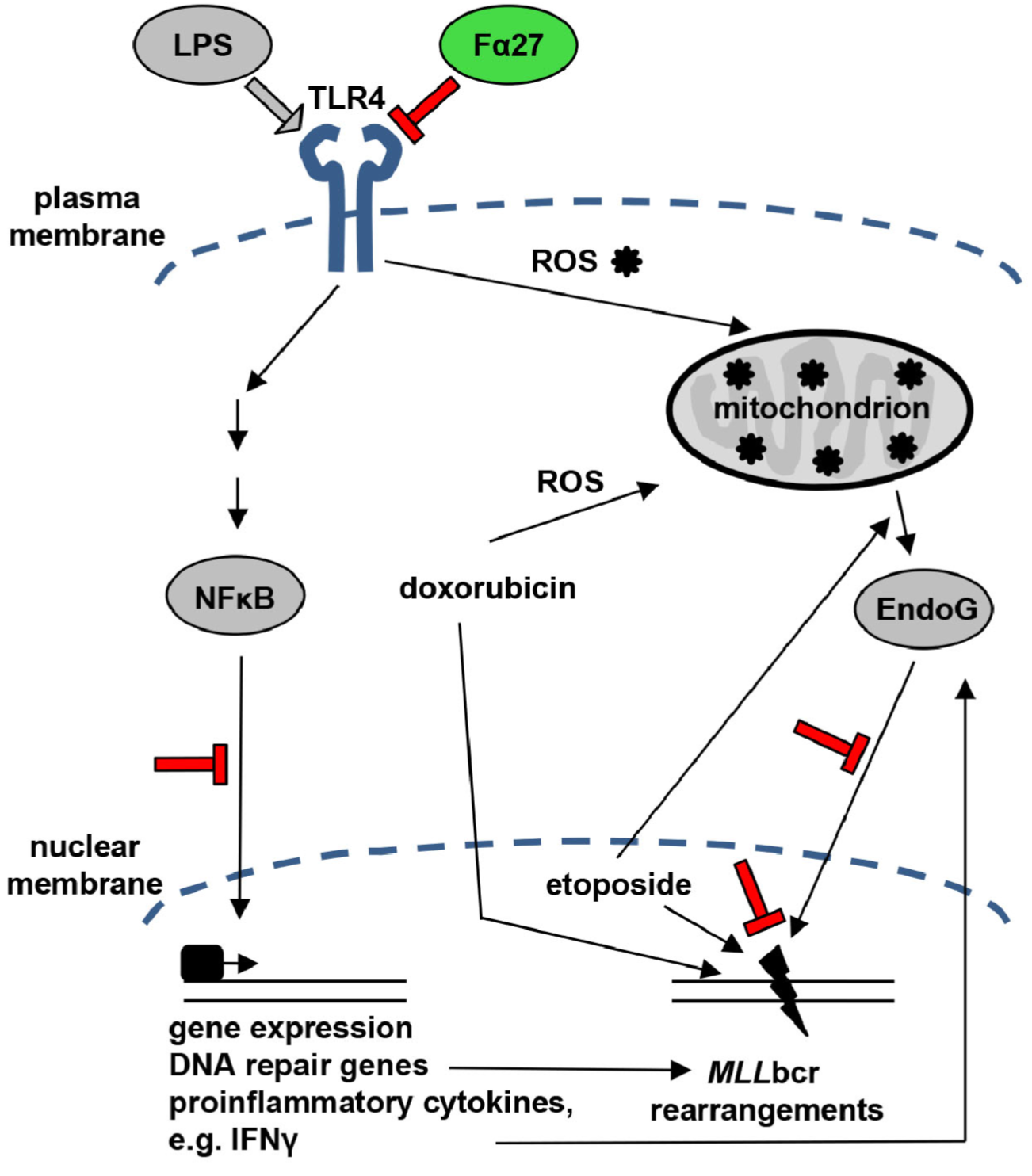
A Fibrinogen Alpha Fragment Mitigates Chemotherapy-Induced MLL Rearrangements
Rearrangements in the Mixed Lineage Leukemia breakpoint cluster region (MLLbcr) are frequently involved in therapy-induced leukemia, a severe side effect of anti-cancer therapies. Previous work unraveled Endonuclease G as the critical nuclease causing initial breakage in the MLLbcr in response to different types of chemotherapeutic treatment. To identify peptides protecting against therapy-induced leukemia, we screened a hemofiltrate-derived peptide library by use of an enhanced green fluorescent protein (EGFP)-based chromosomal reporter of MLLbcr rearrangements. Chromatographic purification of one active fraction and subsequent mass spectrometry allowed to isolate a C-terminal 27-mer of fibrinogen α encompassing amino acids 603 to 629. The chemically synthesized peptide, termed Fα27, inhibited MLLbcr rearrangements in immortalized hematopoietic cells following treatment with the cytostatics etoposide or doxorubicin. We also provide evidence for protection of primary human hematopoietic stem and progenitor cells from therapy-induced MLLbcr breakage. Of note, fibrinogen has been described to activate toll-like receptor 4 (TLR4). Dissecting the Fα27 mode-of action revealed association of the peptide with TLR4 in an antagonistic fashion affecting downstream NFκB signaling and pro-inflammatory cytokine production. In conclusion, we identified a hemofiltrate-derived peptide inhibitor of the genome destabilizing events causing secondary leukemia in patients undergoing chemotherapy.
https://www.frontiersin.org/article/10.3389/fonc.2021.689063
Single-molecule imaging of the transcription factor SRF reveals prolonged chromatin-binding kinetics upon cell stimulation
Serum response factor (SRF) mediates immediate early gene (IEG) and cytoskeletal gene expression programs in almost any cell type. So far, SRF transcriptional dynamics have not been investigated at single-molecule resolution. We provide a study of single Halo-tagged SRF molecules in fibroblasts and primary neurons. In both cell types, individual binding events of SRF molecules segregated into three chromatin residence time regimes, short, intermediate, and long binding, indicating a cell type-independent SRF property. The chromatin residence time of the long bound fraction was up to 1 min in quiescent cells and significantly increased upon stimulation. Stimulation also enhanced the long bound SRF fraction at specific timepoints (20 and 60 min) in both cell types. These peaks correlated with activation of the SRF cofactors MRTF-A and MRTF-B (myocardin-related transcription factors). Interference with signaling pathways and cofactors demonstrated modulation of SRF chromatin occupancy by actin signaling, MAP kinases, and MRTFs.
doi.org/10.1073/pnas.1812734116
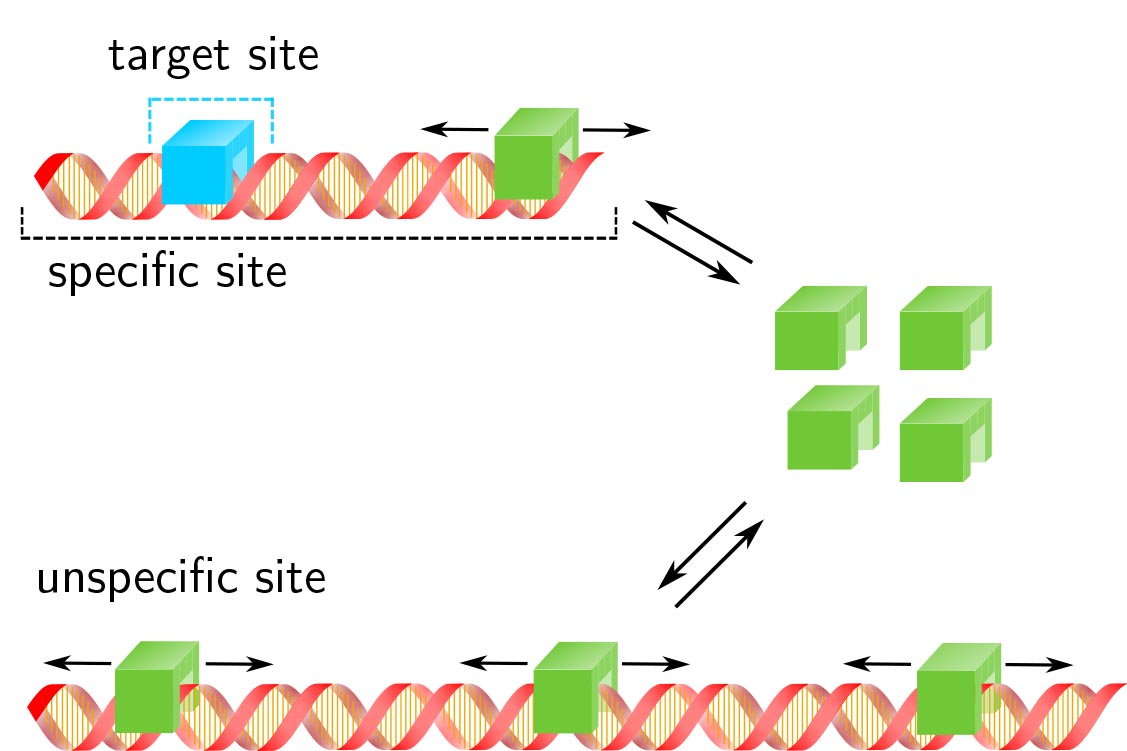
Transcription factor target site search and occupation in a background of unspecific sites
Response time and transcription level are vital parameters of gene regulation. They depend on how fast transcription factors (TFs) find and how efficient they occupy their specific target sites. It is well known that target site search is accelerated by TF binding to and sliding along unspecific DNA and that unspe- cific associations alter the occupation frequency of a gene. However, whether target site search time and occupation frequency can be optimized simultaneously is mostly unclear. We developed a transparent and intuitively accessible state-based formalism to calculate search times to target sites on and occupa- tion frequencies of promoters of arbitrary state structure. Our formalism is based on dissociation rate constants experimentally accessible in live cell experiments. To demonstrate our approach, we consider promoters activated by a single TF, by two coactivators or in the presence of a competitive inhibitor. We find that target site search time and promoter occupancy differentially vary with the unspecific dissocia- tion rate constant. Both parameters can be harmonized by adjusting the specific dissociation rate constant of the TF. However, while measured DNA residence times of various eukaryotic TFs correspond to a fast search time, the occupation frequencies of target sites are generally low. Cells might tolerate low target site occupancies as they enable timely gene regulation in response to a changing environment.
https://doi.org/10.1016/j.jtbi.2018.05.037
DNA residence time is a regulatory factor of transcription repression
Transcription comprises a highly regulated sequence of intrinsically stochastic processes, resulting in bursts of transcription intermitted by quiescence. In transcription activation or repression, a transcription factor binds dynamically to DNA, with a residence time unique to each factor. Whether the DNA residence time is important in the transcription process is unclear. Here, we designed a series of transcription repressors differing in their DNA residence time by utilizing the modular DNA binding domain of transcription activator-like effectors (TALEs) and varying the number of nucleotide-recognizing repeat domains. We characterized the DNA residence times of our repressors in living cells using single molecule tracking. The residence times depended non-linearly on the number of repeat domains and differed by more than a factor of six. The factors provoked a residence time-dependent decrease in transcript level of the glucocorticoid receptor-activated gene SGK1. Down regulation of transcription was due to a lower burst frequency in the presence of long binding repressors and is in accordance with a model of competitive inhibition of endogenous activator binding. Our single molecule experiments reveal transcription factor DNA residence time as a regulatory factor controlling transcription repression and establish TALE-DNA binding domains as tools for the temporal dissection of transcription regulation.
https://doi.org/10.1093/nar/gkx728

Live cell single-molecule imaging of transcription factor binding to DNA
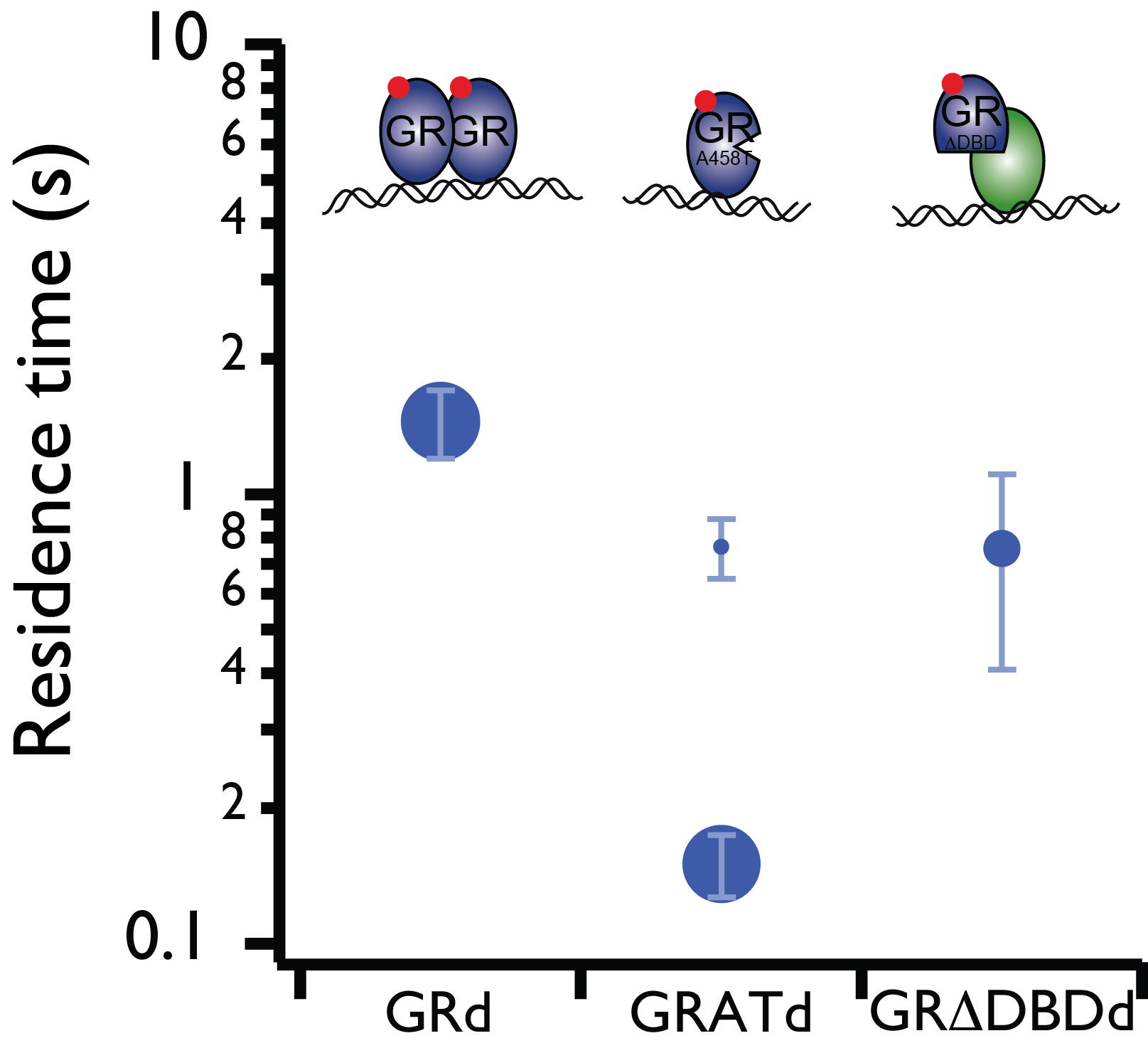
Imaging single fluorescent proteins in living mammalian cells is challenged by out-of-focus fluorescence excitation. To reduce out-of-focus fluorescence we developed reflected light-sheet microscopy (RLSM), a fluorescence microscopy method allowing selective plane illumination throughout the nuclei of living mammalian cells. We demonstrated the single-molecule sensitivity of RLSM by measuring the DNA-bound fraction of glucocorticoid receptor (GR) and determining the residence times on DNA of various oligomerization states and mutants of GR and estrogen receptor-α (ER), which permitted us to resolve different modes of DNA binding of GR. We demonstrated two-color single-molecule imaging by observing the spatiotemporal colocalization of two different protein pairs. Our single-molecule measurements and statistical analysis revealed dynamic properties of transcription factors.
doi: 10.1038/nmeth.2411
Super-resolved spatial organization of RNA polymerase II
Superresolution microscopy based on single-molecule centroid determination has been widely applied to cellular imaging in recent years. However, quantitative imaging of the mammalian nucleus has been challenging due to the lack of 3D optical sectioning methods for normal-sized cells, as well as the inability to accurately count the absolute copy numbers of biomolecules in highly dense structures. Using reflected light-sheet superresolution microscopy, we probed the spatial organization of transcription by RNA polymerase II (RNAP II) molecules and quantified their global extent of clustering inside the mammalian nucleus. Spatiotemporal clustering analysis that leverages on the blinking photophysics of specific organic dyes showed that the majority (>70%) of the transcription foci originate from single RNAP II molecules, and no significant clustering between RNAP II molecules was detected within the length scale of the reported diameter of “transcription factories.” The methods developed in our study pave the way for quantitative mapping and stoichiometric characterization of key biomolecular species deep inside mammalian cells.
doi: 10.1073/pnas.1318496111