Single molecule biophysics in living organisms
Current research:
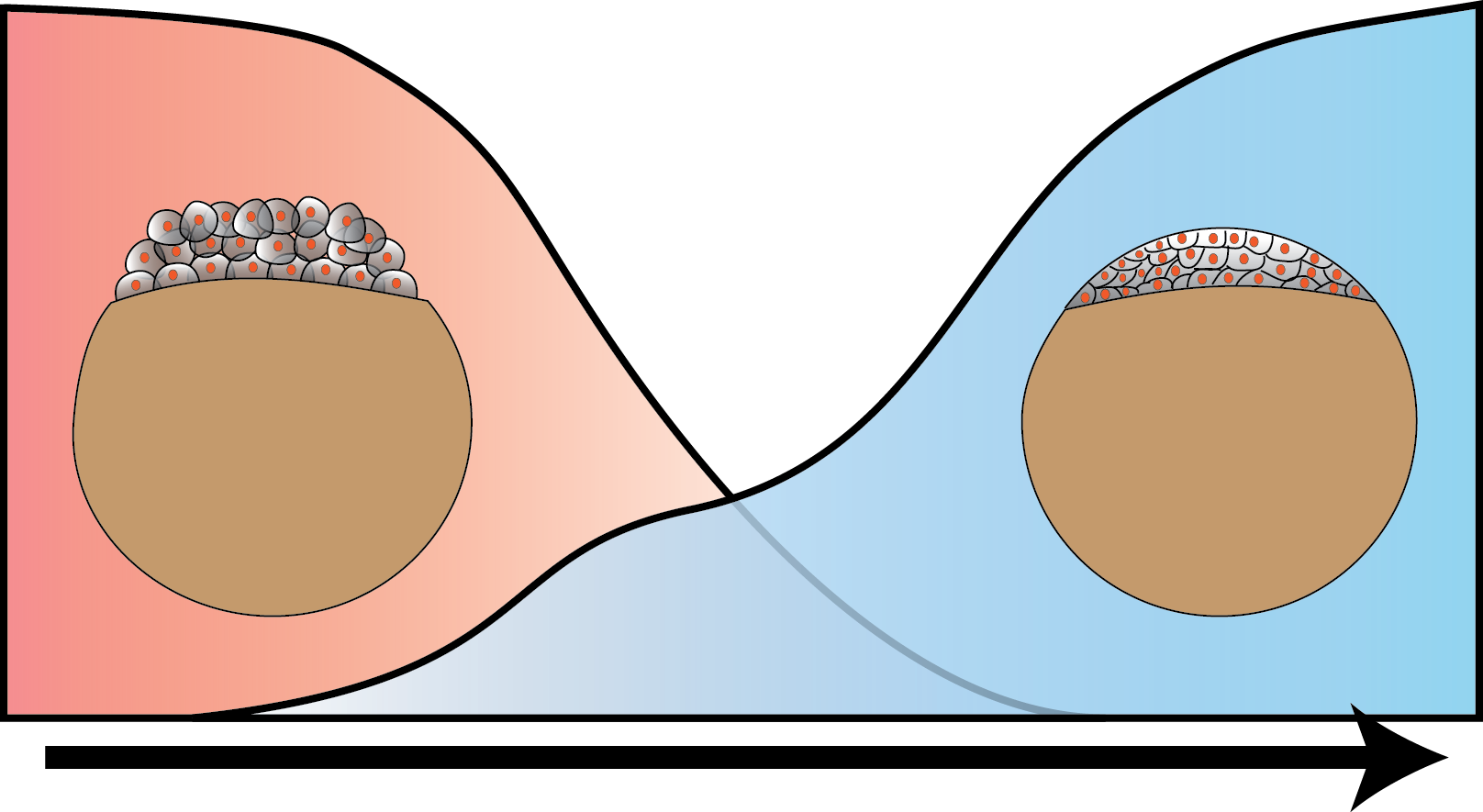
In higher organisms, the early phase of embryonic development is driven by maternally inherited protein and mRNA. After a species-dependent number of cell cycles the zygotic genome is activated and its genes are transcribed (zygotic genome activation, ZGA). We investigate the molecular mechanisms underlying ZGA in live developing Zebrafish embryos at the single molecule level by reflected light-sheet microscopy. Our time and space resolved kinetic measurements allow us to build quantitative models of ZGA.
Published Projects:
→ Embryo development
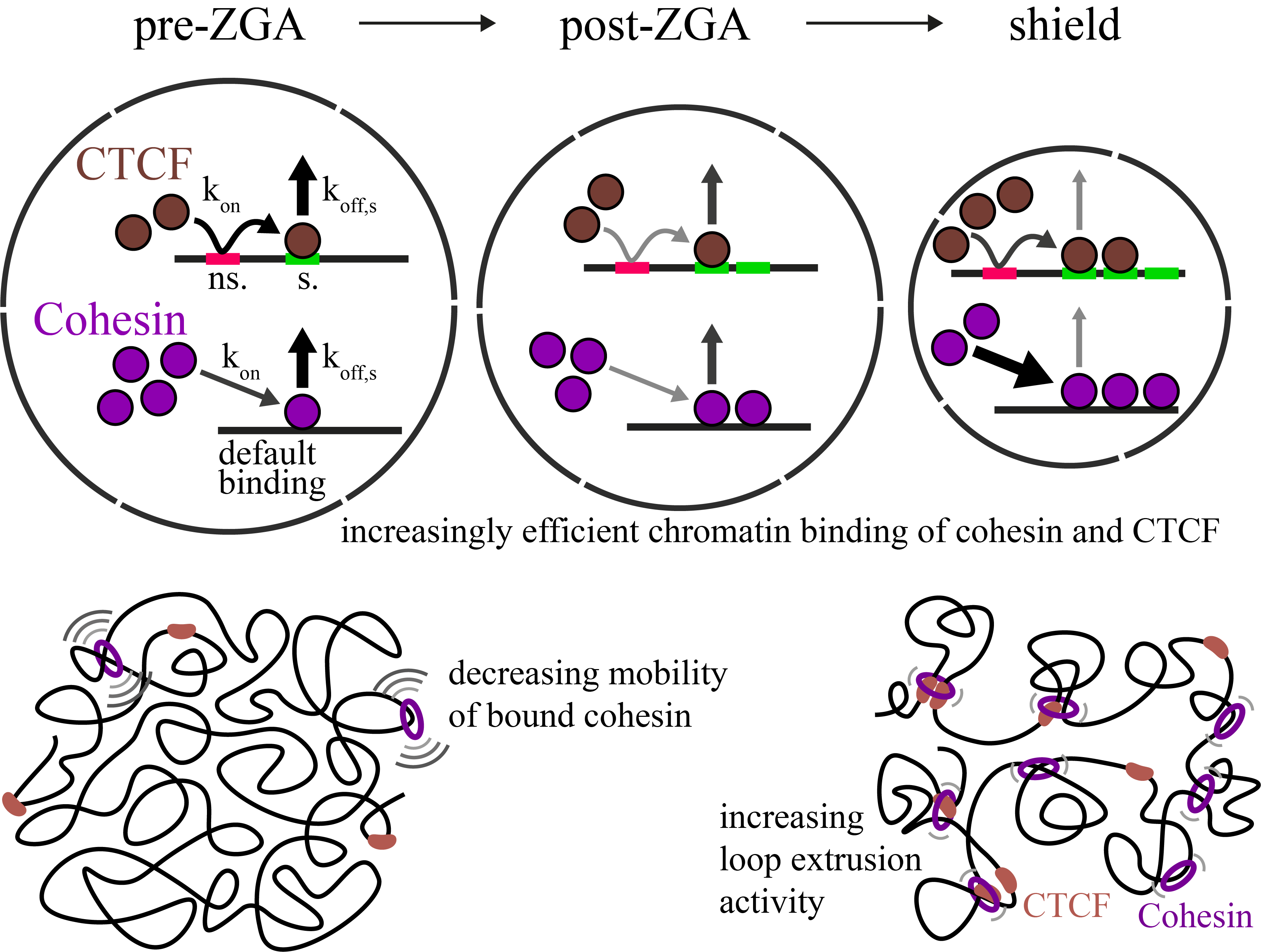
Increasingly efficient chromatin binding of cohesin and CTCF supports chromatin architecture formation during zebrafish embryogenesis
The three-dimensional folding of chromosomes is essential for nuclear functions such as DNA replication and gene regulation. The emergence of chromatin architecture is thus an important process during embryogenesis. To shed light on the molecular and kinetic underpinnings of chromatin architecture formation, we characterized biophysical properties of cohesin and CTCF binding to chromatin and their changes upon cofactor depletion using single-molecule imaging in live developing zebrafish embryos. We found that chromatin-bound fractions of both cohesin and CTCF increased significantly between the 1000-cell and shield stages, which we could explain through changes in both their association and dissociation rates. Moreover, increasing binding of cohesin restricted chromatin motion, potentially via loop extrusion, and showed distinct stage-dependent nuclear distribution. Polymer simulations with experimentally derived parameters recapitulated the experimentally observed gradual emergence of chromatin architecture. Our findings suggest a kinetic framework of chromatin architecture formation during zebrafish embryogenesis.
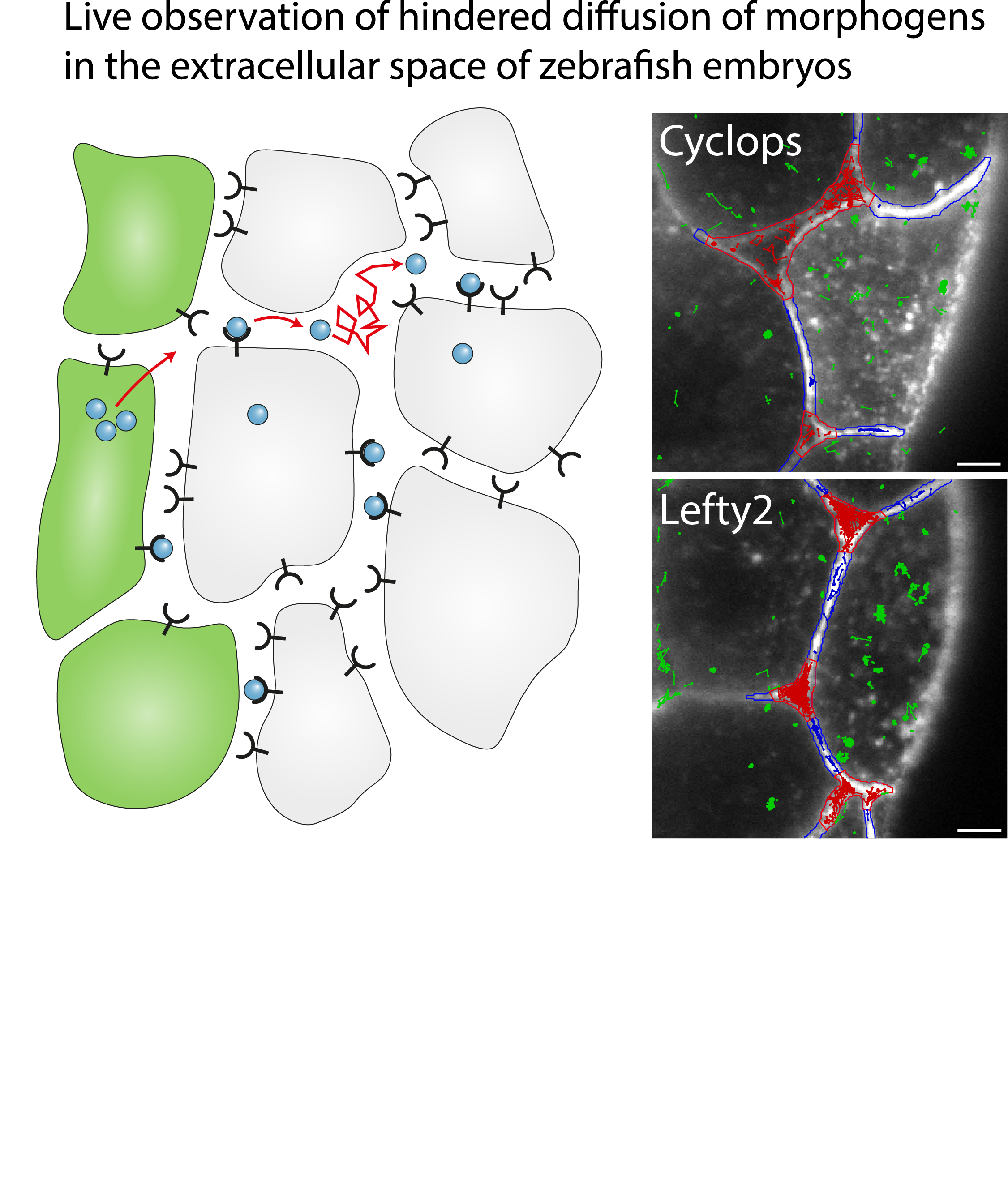
Single-molecule tracking of Nodal and Lefty in live zebrafish embryos supports hindered diffusion model
The hindered diffusion model postulates that the movement of a signaling molecule through an embryo is affected by tissue geometry and binding-mediated hindrance, but these effects have not been directly demonstrated in vivo. Here, we visualize extracellular movement and binding of individual molecules of the activator-inhibitor signaling pair Nodal and Lefty in live developing zebrafish embryos using reflected light-sheet microscopy. We observe that diffusion coefficients of molecules are high in extracellular cavities, whereas mobility is reduced and bound fractions are high within cell-cell interfaces. Counterintuitively, molecules nevertheless accumulate in cavities, which we attribute to the geometry of the extracellular space by agent-based simulations. We further find that Nodal has a larger bound fraction than Lefty and shows a binding time of tens of seconds. Together, our measurements and simulations provide direct support for the hindered diffusion model and yield insights into the nanometer-to-micrometer-scale mechanisms that lead to macroscopic signal dispersal.
https://doi.org/10.1038/s41467-022-33704-z
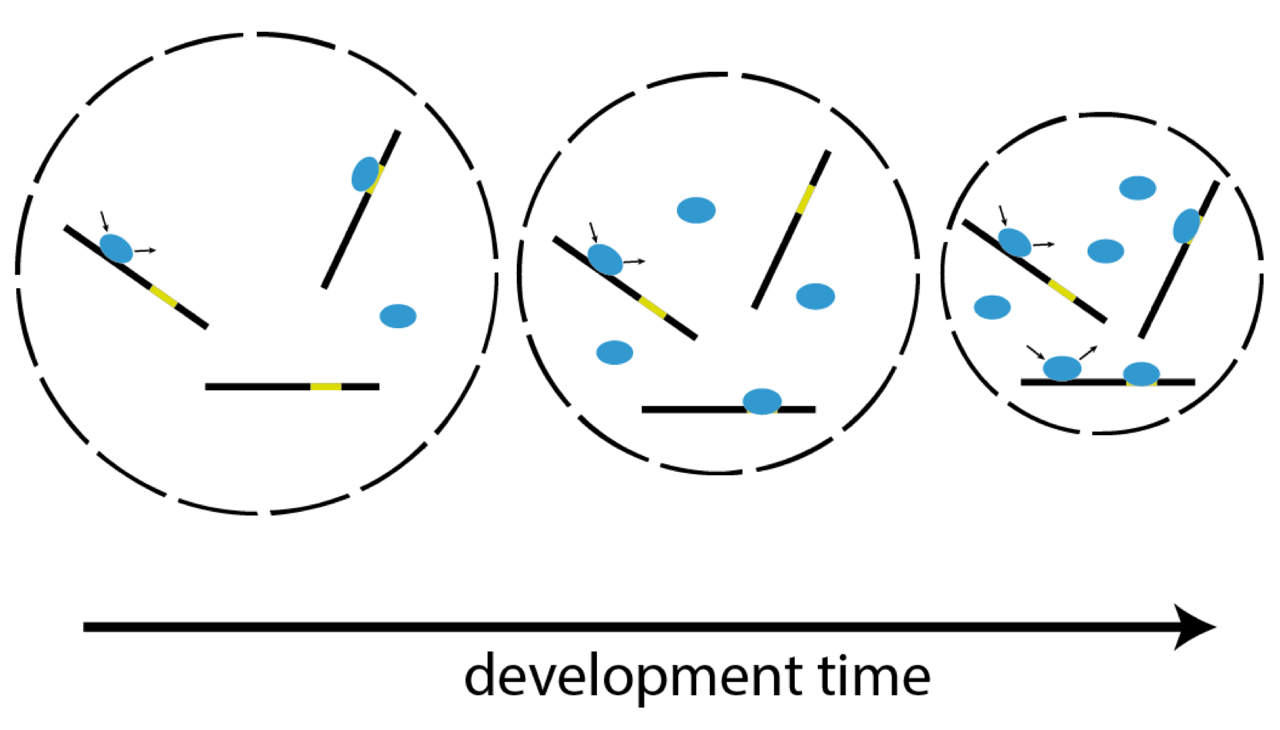
Single-molecule imaging correlates decreasing nuclear volume with increasing TF-chromatin associations during zebrafish development
Zygotic genome activation (ZGA), the onset of transcription after initial quiescence, is a major developmental step in many species, which occurs after ten cell divisions in zebrafish embryos. How transcription factor (TF)-chromatin interactions evolve during early development to support ZGA is largely unknown. We establish single molecule tracking in live developing zebrafish embryos using reflected light-sheet microscopy to visualize two fluorescently labeled TF species, mEos2-TBP and mEos2-Sox19b. We further develop a data acquisition and analysis scheme to extract quantitative information on binding kinetics and bound fractions during fast cell cycles. The chromatin-bound fraction of both TFs increases during early development, as expected from a physical model of TF-chromatin interactions including a decreasing nuclear volume and increasing DNA accessibility. For Sox19b, data suggests the increase is mainly due to the shrinking nucleus. Our single molecule approach provides quantitative insight into changes of TF-chromatin associations during the developmental period embracing ZGA.
https://www.nature.com/articles/s41467-018-07731-8
