Weidinger lab - research
All animals have evolved strategies to deal with damage due to injury or disease, but the ability to regenerate lost or damaged organs and appendages varies greatly in different species. Unfortunately, humans and other mammals are quite poor at regenerating, while other vertebrates, like salamanders and fish, can efficiently re-grow lost limbs/fins and regenerate many internal organs including their hearts. Currently it is a mystery why mammals can’t do what these lower vertebrates achieve. We hope that elucidating the mechanisms that zebrafish use to regulate regeneration will one day result in therapies aimed at activating regenerative potential in human organs.
Mechanisms of zebrafish heart regeneration
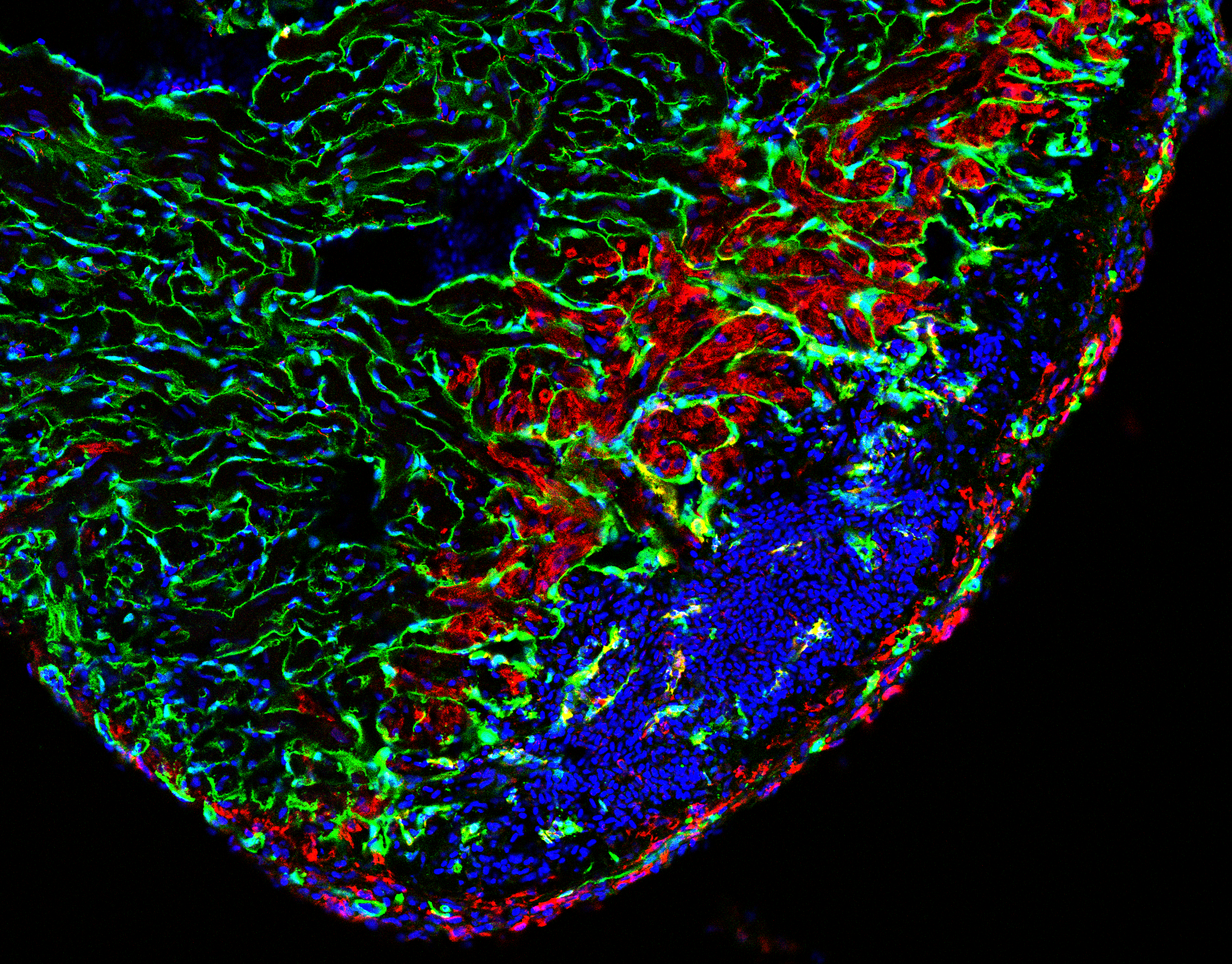

Heart damage, usually caused by infarction, is a leading cause of death in humans. After cardiomyocyte loss the damaged part of the myocardium in humans undergoes extensive scarring and fibrosis, and the cardiomyocytes lost to injury cannot be replaced. Thus, infarction results in permanent damage to the heart. In contrast, zebrafish are able to regenerate their hearts after injury without permanent scarring. Importantly, cardiomyocytes efficiently proliferate to restore to lost muscle tissue. We have found that injuries causing necrotic death of the myocardium, which are similar to human heart infarction, are efficiently and completely regenerated.
Regeneration involves the proliferation of differentiated cardiomyocytes and the coordinated growth of myocardium, endocardium and epicardium. We found that BMP signaling is essential for heart regeneration. Interestingly, it promotes cardiomyocyte proliferation during regeneration, but is not required for physiological cardiomyocyte proliferation. Surprisingly, we discovered that proliferating cardiomyocytes experience replication stress, which is a leading cause of declining tissue repair in aged mammals. Yet, we found that BMP signaling allows zebrafish to overcome this stress and that BMP signaling alleviates replication stress also in human cells. Thus, insights into zebrafish regeneration can result in the discovery of potential anti-aging interventions.
Currently, we further explore the relationship between regeneration and aging and how BMP signaling alleviates replication stress.
Selected first or last author publications of our group related to heart regeneration:
Vasudevarao et al., 2025, Nature Communications
Bertozzi et al. 2022, Dev. Biol.
Bertozzi et al. 2021, Dev. Biol.
Wu et al. 2016, Developmental Cell
Wu and Weidinger 2014, Curr Pathobiol. Reports
Schnabel et al. 2011, PLOS One
Knopf et al. 2010, PNAS
Nemtsas et al. 2010, J Mol Cell Card
Ueno et al. 2007, PNAS
Our work on heart regeneration is embedded in the Collaborative Research Center (SFB) 1506: "Aging at Interfaces".
Mechanisms of zebrafish fin and bone regeneration
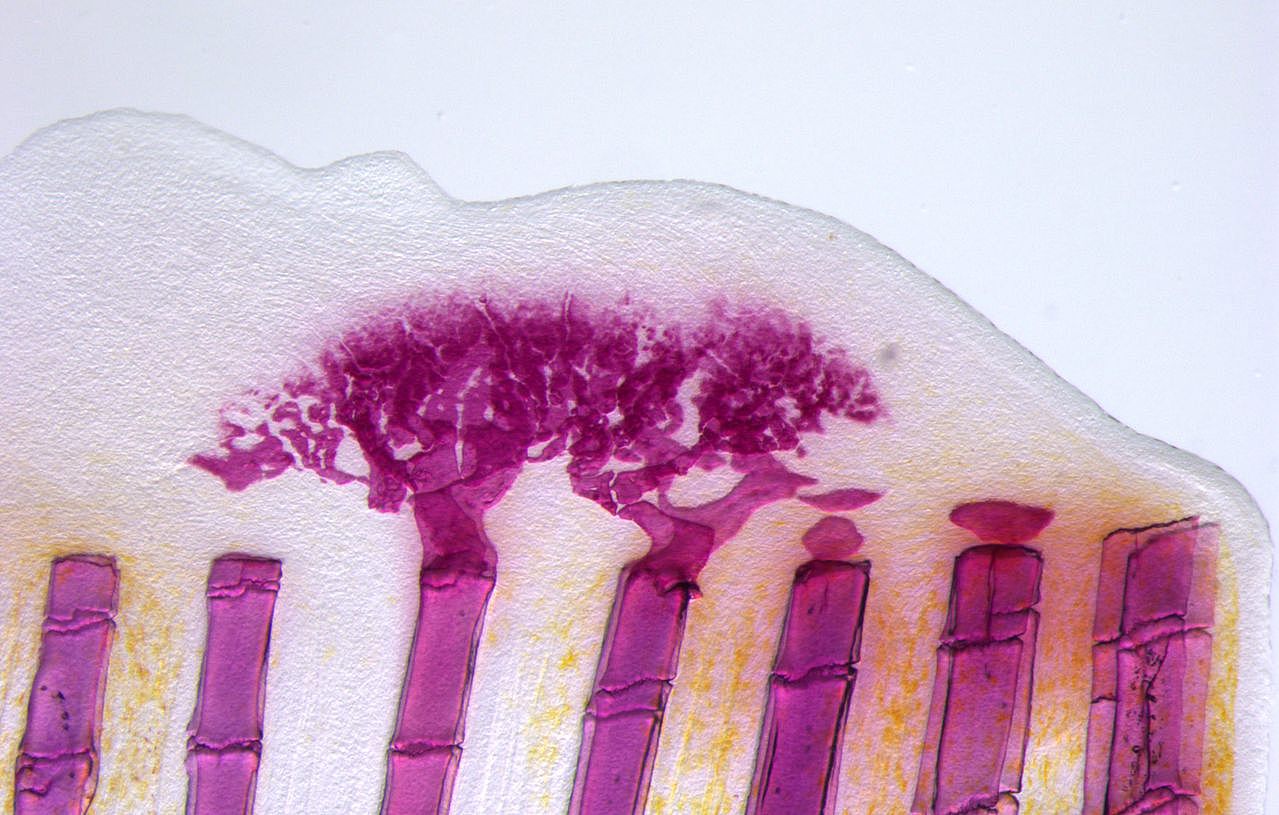
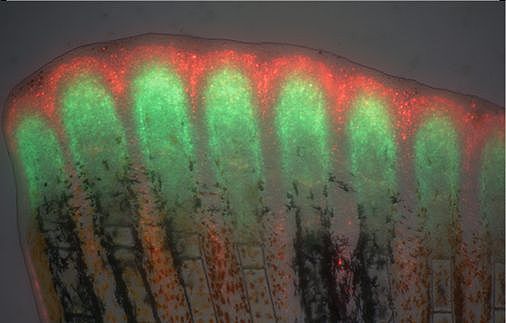
Unlike mammals, zebrafish can completely and repeatedly regenerate lost appendages, that is their fins. This occurs via formation of a population of progenitor cells, the blastema, which contains the precursors of the regenerating tissue. How the blastema forms is a central question in regeneration research. Intriguingly, we have found that mature osteoblasts dedifferentiate and give rise to blastema cells after fin injury. The fact that differentiated cells, and not stem cells, represent important source cells for tissue regeneration has recently been recognized also in several other systems. A major focus of the lab thus has been to understand how osteoblast dedifferentiation is initiated and regulated. We found that NF-kB signaling needs to be downregulated for dedifferentiation to occur. Intriguingly, we also discovered that osteoblast dedifferentiation and another important response of these cells to injury, namely their migration towards the wound, can be uncoupled and are regulated independently. We continue to study molecular mechanisms regulating osteoblast injury responses.
Selected first or communicating author publications of our group related to fin & bone regeneration:
Sehring et al. 2022, elife
Mishra et al. 2020, Developmental Cell
Owlarn et al. 2017, Nature Communications
Sehring & Weidinger 2016, Current Opinion in Genetics & Development
Wehner et al. 2015, JoVE
Wehner et al. 2015, Trends in Genetics
Geurtzen et al 2015, Development
Wehner et al. 2014, Cell Reports
Grotek et al. 2013, Development
Azevedo et al. 2011, PLOS One
Knopf et al. 2011, Developmental Cell
Stoick-Cooper et al. 2007, Genes & Development
Stoick-Cooper et al. 2007, Development

Our work on bone regeneration is embedded in the Collaborative Research Center (SFB) 1149: "Danger Response, Disturbance Factors and Regenerative Potential after Acute Trauma".
Regulation of Wnt signaling pathways
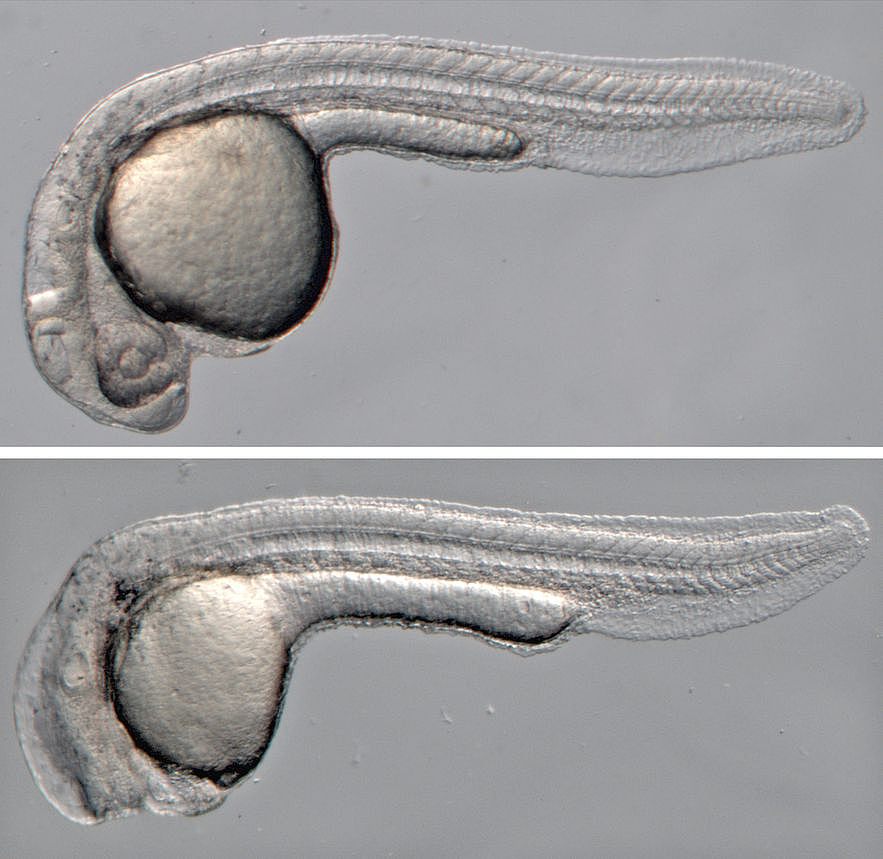
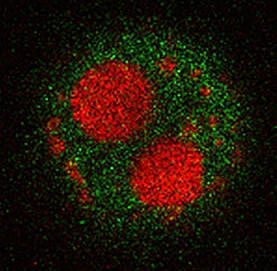
Wnt signaling does not only have essential functions during organ regeneration, but plays important roles in regulating cell fate, cell proliferation, cell polarity and migration in many tissues during embryonic development and in adult homeostasis. Tight regulation of Wnt signaling is essential, in particular of the Wnt/beta-catenin pathway, since misregulation causes disease, most prominently cancer. A long-standing interest of the lab has been to identify regulators of this important pathway.
Currently, we aim to discover Wnt-regulating peptides.
Selected first or last author publications of our group related to the regulation of Wnt signaling:
Özhan et al. 2013, Dev. Cell
Kagermeier-Schenk et al. 2011, Dev. Cell
Nakamura et al. 2007, J Clin Invest.
Our work on Wnt signaling is embedded in the Collaborative Research Center (SFB) 1279: “Exploiting the Human Peptidome for Novel Antimicrobial and Anticancer Agents”.